 Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00637800
Standard Name
UTP14
(small nucleolar ribonucleoprotein complex subunit Utp14)
Aliases
PreTt19783 | 91.m00198
Description
UTP14 Utp14p; Subunit of U3-containing Small Subunit (SSU) processome complex involved in production of 18S rRNA and assembly of small ribosomal subunit; Small-subunit processome, Utp14
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
No Data fetched for Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 GeneMania
GeneMania
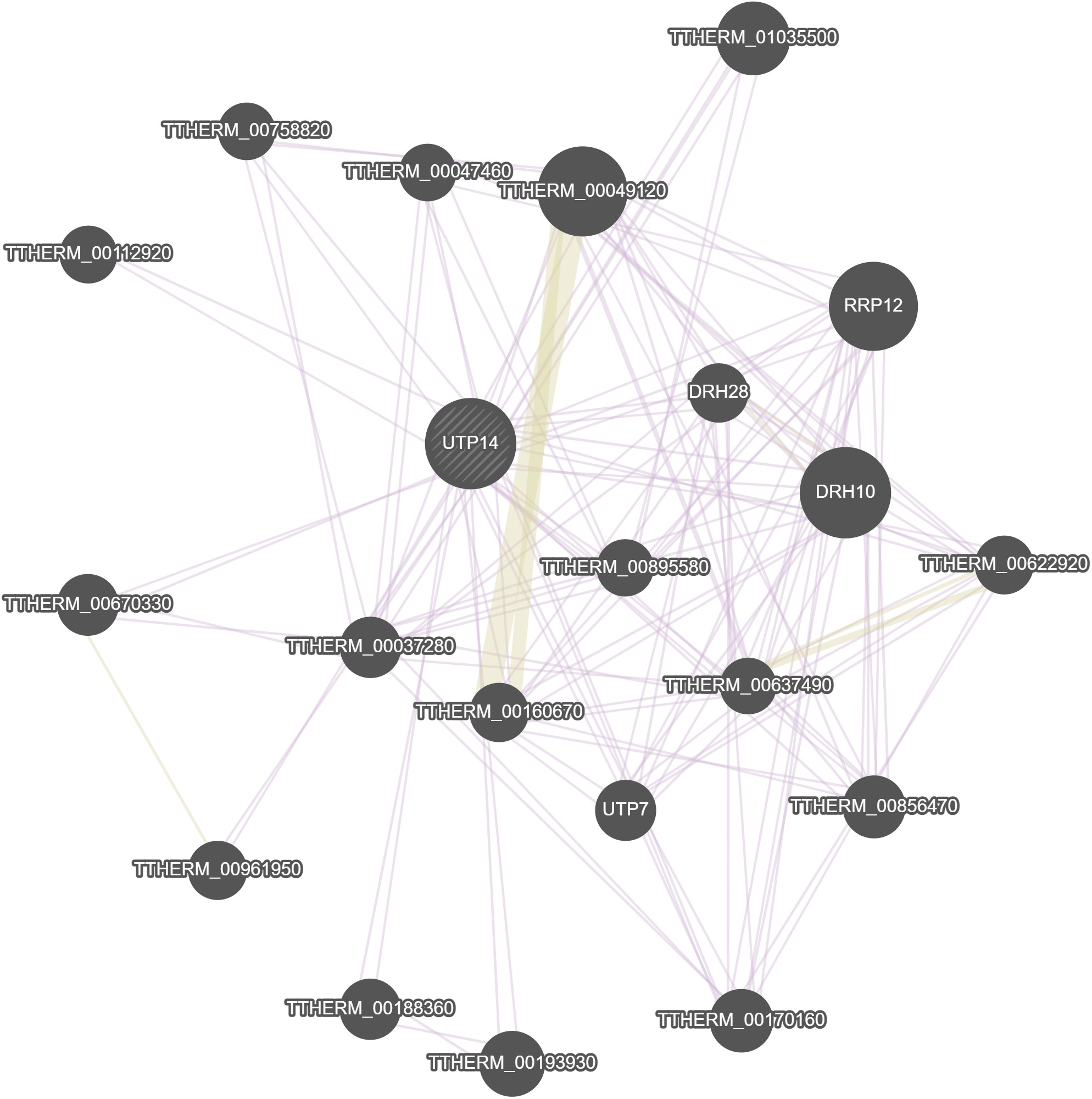
No results were calculated for this gene in GeneMania.
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00637800(coding)
ATGGCTGATAAAAAGAAAATTAAAAAGTAGGATAAAGAAGAGAAGTATAAAAAGCTATTA
CAAGCTATTGACATCAAGCTTGGTGAGGAGAAGGTTGATGAAAAATAAAAGAAAGTAGAA
GATCTCATTGCATCCTAAAAAAAGAATTAATAAGAATAGTAGAAGCCCGAAGAGGTTTTA
ACAATCTCAGGTCTTCTTTCTAAAATATAAGGAAATGATGAACAAGAGAATGCATAAAAT
TTAAAAAAATAATTAGAGGATCTTGAAAAAAGAAGTAAACTTCTTCATGAGCCAGAGTCT
TTCGTCAAATAGGAGAGAGCTGAAAGAAAAATTAATTATAAAATTGTTTAAGATGATGTA
GCTAAATGGATTCCTATTATTAAAAGGGCTAGAGAGGCTGATCATTTAGATTTTACACAA
AAGACTTAAGATAATTTGAGAATAAATGCAATTTCTGGAGATGAATTAAAAAGAAATAAA
ATTGCCATGAGCATTGAATAAAAATTATAACAATTAAATCTTAAGACTGAAAAGTAGATT
AAAGCTAAGGAGATAGAATTATTAGAAAAATTAGATCCTTAGGAAGCTAAAAGAAGAATT
AATGAAATGAATAAACAAAAAATGTTACTCTTCTAACAAGAAATTAAAAACAAAAGAGTA
GCTAAAATTAAGAGTAAGCTCTACCATAGAATTAAAAAGAAACAAAAACTCAGAGCTCAA
TTAAAAGAATATGAAAACATGGATGAAGAAGCAAGATTGTAATAGTAATAGGAATTATTA
GAAGAAAGAGCCAAAGAAAGAATTGGTATGAGACATAAAACTAAATCTAAACACATTTAA
TAAATGTTGAAGTATGGTGATAAGAAAACCTACTAATAGTCATTAAATGATATTAATCAT
TAGAGATAAGCTTTACTTAAAAAAACATAAGACATAGAAGTTAATTCAGATGATTTAGAT
TATGACTAACTCAATGATGAAGAATTTAAAGAACAAGCTATAAAAGACTTATAAAATGAG
CTTAAAGGAGGGGATGATGATGAATAGCAAGAAGAGCTAACTGGTATTAAATTCATTGAT
GATCTTCATAATTAAAAAAAGAATCAACATAGAGATGAAGCTCGTAAAATCATTGCTTAA
TTGTAAGATCCTGATGCTTTTGATAAAGAGATTTTGTCAGAAGATGATTTAATTGAAAGC
GATGATGAAGAAGGATAAAAGAAAAAAGCAGATAAGTTGAATGCAAAGAGAAAAGCTGAT
TAGGAAATTTTAGAAAAGGAAGAAGAATTCACAGGAAGAAAAATTTACAAAAAATCTGAA
CCTGAAGAGAAGTAAACAGAATTAGAAATCGAGATGGAGGAAATGAAATAAGCTAAAAAA
AAGAAAAGATTGAATGCTCTCGACATTGTAAAGGATCATATTAAATCTTAAAAGGAATAA
GAAGAAAACGAATAGGAAATTGAAAATGAATAATCTAAGAGTATTTCATAAAACGGAACA
AATAATAAGAAATAAGAAATATCTCAATAATCTACTATATAAGAAAAACTCAAAGAAGCA
TCAAAAAAAGTAGCAGCAACAACAATTAATGAAGAAAATTTGACTGATGGTTTGAACGTT
AATAAATTGAAAGAAAAATTCAAAAAAAATGCATAAACAGAGTTCAGTAAAAAGGAACTT
GAAGCATTCAACATAGAAAACAAAGAAGAAATCTACAAGGATTTATCTAATTTTAATGTT
TTAGTAGATGATGATAATGAGTAATAGTTTTTGGCAGAAAAGTATGAAGATTACGAAAAA
GACTTACCATAAGAGAAAAAGCCTTTATAAGGTTGGGGACATTGGGTTGGATAAGGTATT
GAATAAAAGAATTTACCTACTGCAGAAGAATTAGCAAGACAAAAGGCTGCTAAAATAAAT
TTACTCAAAAAGTAGAGAGCCGATGGCAGCATGGATAATGTTATATTGAACGAAAAGAGA
AATAAATTATTTACTTAGCATTTAGTCAAAGAACTTCCTCACCCATACAAAAACAAAGAA
CAATTCGAATACTTACACAATGCACCAATAGGTTCAGAATGGACAACTATGCGTAGTCAT
GTCAACTTAATCAAACCCAAAATAAAAACAAAGGCAGGAAACATTATTCAACCAGCTACC
GCTCCTAAATCCTTCTAATAATCTTGA
>TTHERM_00637800(gene)
AATTAAGCATGCAAATGACTATTATTACTACTTGCTACATATGCATATTTGTATAGATTT
GATATTCAATCAAAAATAAAACAAACAAGAATTTATTTTATAAGCACAACTATATTATTT
TAAGTAGATTGCTATTAACCATGGCTGATAAAAAGAAAATTAAAAAGTAGGATAAAGAAG
AGAAGTATAAAAAGCTATTACAAGCTATTGACATCAAGCTTGGTGAGGAGAAGGTTGATG
AAAAATAAAAGAAAGTAGAAGATCTCATTGCATCCTAAAAAAAGAATTAATAAGAATAGT
AGAAGCCCGAAGAGGTTTTAACAATCTCAGGTCTTCTTTCTAAAATATAAGGAAATGATG
AACAAGAGAATGCATAAAATTTAAAAAAATAATTAGAGGATCTTGAAAAAAGAAGTAAAC
TTCTTCATGAGCCAGAGTCTTTCGTCAAATAGGAGAGAGCTGAAAGAAAAATTAATTATA
AAATTGTTTAAGATGATGTAGCTAAATGGATTCCTATTATTAAAAGGGCTAGAGAGGCTG
ATCATTTAGATTTTACACAAAAGACTTAAGATAATTTGAGAATAAATGCAATTTCTGGAG
ATGAATTAAAAAGAAATAAAATTGCCATGAGCATTGAATAAAAATTATAACAATTAAATC
TTAAGACTGAAAAGTAGATTAAAGCTAAGGAGATAGAATTATTAGAAAAATTAGATCCTT
AGGAAGCTAAAAGAAGAATTAATGAAATGAATAAACAAAAAATGTTACTCTTCTAACAAG
AAATTAAAAACAAAAGAGTAGCTAAAATTAAGAGTAAGCTCTACCATAGAATTAAAAAGA
AACAAAAACTCAGAGCTCAATTAAAAGAATATGAAAACATGGATGAAGAAGCAAGATTGT
AATAGTAATAGGAATTATTAGAAGAAAGAGCCAAAGAAAGAATTGGTATGAGACATAAAA
CTAAATCTAAACACATTTAATAAATGTTGAAGTATGGTGATAAGAAAACCTACTAATAGT
CATTAAATGATATTAATCATTAGAGATAAGCTTTACTTAAAAAAACATAAGACATAGAAG
TTAATTCAGATGATTTAGATTATGACTAACTCAATGATGAAGAATTTAAAGAACAAGCTA
TAAAAGACTTATAAAATGAGCTTAAAGGAGGGGATGATGATGAATAGCAAGAAGAGCTAA
CTGGTATTAAATTCATTGATGATCTTCATAATTAAAAAAAGAATCAACATAGAGATGAAG
CTCGTAAAATCATTGCTTAATTGTAAGATCCTGATGCTTTTGATAAAGAGATTTTGTCAG
AAGATGATTTAATTGAAAGCGATGATGAAGAAGGATAAAAGAAAAAAGCAGATAAGTTGA
ATGCAAAGAGAAAAGCTGATTAGGAAATTTTAGAAAAGGAAGAAGAATTCACAGGAAGAA
AAATTTACAAAAAATCTGAACCTGAAGAGAAGTAAACAGAATTAGAAATCGAGATGGAGG
AAATGAAATAAGCTAAAAAAAAGAAAAGATTGAATGCTCTCGACATTGTAAAGGATCATA
TTAAATCTTAAAAGGAATAAGAAGAAAACGAATAGGAAATTGAAAATGAATAATCTAAGA
GTATTTCATAAAACGGAACAAATAATAAGAAATAAGAAATATCTCAATAATCTACTATAT
AAGAAAAACTCAAAGAAGCATCAAAAAAAGTAGCAGCAACAACAATTAATGAAGAAAATT
TGACTGATGGTTTGAACGTTAATAAATTGAAAGAAAAATTCAAAAAAAATGCATAAACAG
AGTTCAGTAAAAAGGAACTTGAAGCATTCAACATAGAAAACAAAGAAGAAATCTACAAGG
ATTTATCTAATTTTAATGTTTTAGTAGATGATGATAATGAGTAATAGTTTTTGGCAGAAA
AGTATGAAGATTACGAAAAAGACTTACCATAAGAGAAAAAGCCTTTATAAGGTTGGGGAC
ATTGGGTTGGATAAGGTATTGAATAAAAGAATTTACCTACTGCAGAAGAATTAGCAAGAC
AAAAGGCTGCTAAAATAAATTTACTCAAAAAGTAGAGAGCCGATGGCAGCATGGATAATG
TTATATTGAACGAAAAGAGAAATAAATTATTTACTTAGCATTTAGTCAAAGAACTTCCTC
ACCCATACAAAAACAAAGAACAATTCGAATACTTACACAATGCACCAATAGGTTCAGAAT
GGACAACTATGCGTAGTCATGTCAACTTAATCAAACCCAAAATAAAAACAAAGGCAGGAA
ACATTATTCAACCAGCTACCGCTCCTAAATCCTTCTAATAATCTTGATAAAAATAGAGAT
TAAGTTTTATAGATAAAATAGAAACCAACTAAATTATAATTTTCATCTAAATGCATTCTA
ATAATTCAAAAAATAAATATTGTATTTTTATTTATTTAGATAAAAACGTGTATAAAATTA
TCTATTCATCAAT
>TTHERM_00637800(protein)
MADKKKIKKQDKEEKYKKLLQAIDIKLGEEKVDEKQKKVEDLIASQKKNQQEQQKPEEVL
TISGLLSKIQGNDEQENAQNLKKQLEDLEKRSKLLHEPESFVKQERAERKINYKIVQDDV
AKWIPIIKRAREADHLDFTQKTQDNLRINAISGDELKRNKIAMSIEQKLQQLNLKTEKQI
KAKEIELLEKLDPQEAKRRINEMNKQKMLLFQQEIKNKRVAKIKSKLYHRIKKKQKLRAQ
LKEYENMDEEARLQQQQELLEERAKERIGMRHKTKSKHIQQMLKYGDKKTYQQSLNDINH
QRQALLKKTQDIEVNSDDLDYDQLNDEEFKEQAIKDLQNELKGGDDDEQQEELTGIKFID
DLHNQKKNQHRDEARKIIAQLQDPDAFDKEILSEDDLIESDDEEGQKKKADKLNAKRKAD
QEILEKEEEFTGRKIYKKSEPEEKQTELEIEMEEMKQAKKKKRLNALDIVKDHIKSQKEQ
EENEQEIENEQSKSISQNGTNNKKQEISQQSTIQEKLKEASKKVAATTINEENLTDGLNV
NKLKEKFKKNAQTEFSKKELEAFNIENKEEIYKDLSNFNVLVDDDNEQQFLAEKYEDYEK
DLPQEKKPLQGWGHWVGQGIEQKNLPTAEELARQKAAKINLLKKQRADGSMDNVILNEKR
NKLFTQHLVKELPHPYKNKEQFEYLHNAPIGSEWTTMRSHVNLIKPKIKTKAGNIIQPAT
APKSFQQS
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences



