Gene Model Identifier
TTHERM_00657270
Standard Name
RRS1
(arginyl-tRNA synthetase 1)
Aliases
PreTt02111 | 97.m00097 | 3724.m00005
Description
RRS1 arginyl-tRNA synthetase protein; arginyl-tRNA synthetase family protein; Arginine-tRNA ligase
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00657270(coding)
ATGGAGTCTAAAGTTTTAAAAAGTATGATGAAAATCGCATCTCAACACACATTAAACGTT
GTTAACAATGCTTTGAAGCCCTACAATGTTTAATCTCCTCCTATCAAGATTTAACCTGTT
TCTAACTTAGTTGATGTTGATTACAACACCCCAACTCTCTTGCAAGTCTTCAACCAAAAT
AAAGATAAGAAAACAAAGCAAGCTTTTGGTTTGGCAGACATTAAGGCATTCTGTCATAAG
GTTATTGAATCATTCAATCCTGAAGATAAGGACTGTGCTATCGAAAAGTTAGGTGTTACT
GACTAAGGTTTTTTGACTATTAAGTTAAAATAATAAGTTATTGAAGATGAAATTAACAAT
ATTTTAAAGAATGGTTTGAACTACCCCACTGAAAAGAAACAAAAGATTGCCGTTGATTTC
TCATCTCCTAATATTGCCAAAGAAATGCACGTTGGTCACTTGCGTTCAACTATTATTGGT
GAATCTATTTGCCGTATTTTTGAATTCTAAAACCACGAAGTTCTTCGTATTAACCATTTG
GGTGATTGGGGTACTCAATTCGGTATGCTTATTACTTATATGAAGAGAAAATATCCTAAT
TTCCTCAATGAACCTATTGATTTATCAGATTTAACTATGTTCTACAAGGAATCTAAGAAG
TTATTCGATGAAGATCCTGAATTCAAGAAGACTGCTTAACTTACTGTCGTCGACCTTCAA
AGTGGTGACCCTGAATGTCGCAAGGCTTGGGAAATTCTTTGTGAAATTTCTCGTAGGGAA
TTCACTGAAATCTACAAGCGTTTAGACATTACTTTAGAAGACTTCGGTGAGTCTTTCTAC
AACGATAAAATTCCTGCTGTCATTTAAGACCTCAAGGATGCTGGTATGCTTGTTGAAAGT
GATGGTGCTCAATGTGTTTTCATTGAAAAGTATAAGAACTAACCTCCTCTCTTCATTGTC
AAAAAGGATGGTGGTTACGGATACGATAGTACTGATATGGCCTGCATCAAGTATAGATTA
AAAACTTTGAACTGCAATCGTTTAGTTTACATTACTGATGTTGGATAACAACCTCATTTT
GATTATATTATTGCTGCTGCTGAAAAAATCGGATGGCACGTTCCTCCCACTACTCGTGCT
GAGCATATGGGTTTCGGTATCGTCTAAGGTGAAGACGGTAAGCGTTTCCGTACTCGTTCC
AGTGAAACTGTCAAGCTAAAAGATTTACTTGATGAAGCTAGAGAACGTGCTTTAGAAAAA
ATTAAAGAACGTATTTCAGGTGGTGCTAAAGAAGAAGGTGCTGAAGAAGATGAAGAAGAA
AAAGAAAACAAGGGTAACTACACCAAGCTCTCAGAAGAAGAATATCTTGATGCTGCTGAA
AAGATGGGTATGGCTGCTATTAAATATTACGATTTACGTTAAAACAGAATCAGCAACTAT
AACTTCTCTTATGATAAAATGCTTGACACTAAGGGTAATACTGCTGTCTACTTACTCTAT
TCCTATGCTCGTCTTTGCTCCATCATCCGTAAGAGTGGTGTCTAGTCTGAAGAACTCTGC
AAAATTATCTAAACTTAAGGATTCAAGATTTCTCATCCCCAAGAAAGAATCATTGCTTCC
CTCCTCATAAAGTTCCCTGATGTTCTTGACTAGGTTACTGAAGATTTAGCAATTAATCGT
TTAACTGACTATATGTATGACCTTGCTTGCAAAGTAGCTGAAGGTTATCGTAAATATCAC
ATCAACAATTCTGAAGATAAGCTTACTCGTATCCTTCTCATTGAAGCTACTCGTTAAGTC
CTTGAAAAGTGTTTCTACTTGGTTGGTATCAAGCCCCTTGAAAAAATCTGA
>TTHERM_00657270(gene)
ATAACGAATATAATATTGGTTAATTAATTTGACATTATTGATGAAATTGTCAGTTTTTGG
CGTAATATCTAATCTAAAGCATCTATGTATGTTAAATTAAATTTTAACTATTTTGTGATG
ATAGAAACTTGGGTATATATTTGCATATATTAGATGCACAAATGTTGGGTTTAAAAGTAC
AAAAAATTAGTTGCCAAAAAATAATTATTTAATTAAAAAATAATAAAAGAAGAGAAATTA
TTTGAATATTCTAAAGAAAATAAATAGATACATAGCAACTTTAAATATAAATGGAGTCTA
AAGTTTTAAAAAGTATGATGAAAATCGCATCTCAACACACATTAAACGTTGTTAACAATG
CTTTGAAGCCCTACAATGTTTAATCTCCTCCTATCAAGATTTAACCTGTTTCTAACTTAG
TTGATGTTGATTACAACACCCCAACTCTCTTGCAAGTCTTCAACCAAAATAAAGATAAGA
AAACAAAGCAAGCTTTTGGTTTGGCAGACATTAAGGCATTCTGTCATAAGGTTATTGAAT
CATTCAATCCTGAAGATAAGGACTGTGCTATCGAAAAGTTAGGTGTTACTGACTAAGGTT
TTTTGACTATTAAGTTAAAATAATAAGTTATTGAAGATGAAATTAACAATATTTTAAAGA
ATGGTTTGAACTACCCCACTGAAAAGAAACAAAAGATTGCCGTTGATTTCTCATCTCCTA
ATATTGCCAAAGAAATGCACGTTGGTCACTTGCGTTCAACTATTATTGGTGAATCTATTT
GCCGTATTTTTGAATTCTAAAACCACGAAGTTCTTCGTATTAACCATTTGGGTGATTGGG
GTACTCAATTCGGTATGCTTATTACTTATATGAAGAGAAAATATCCTAATTTCCTCAATG
AACCTATTGATTTATCAGATTTAACTATGTTCTACAAGGAATCTAAGAAGTTATTCGATG
AAGATCCTGAATTCAAGAAGACTGCTTAACTTACTGTCGTCGACCTTCAAAGTGGTGACC
CTGAATGTCGCAAGGCTTGGGAAATTCTTTGTGAAATTTCTCGTAGGGAATTCACTGAAA
TCTACAAGCGTTTAGACATTACTTTAGAAGACTTCGGTGAGTCTTTCTACAACGATAAAA
TTCCTGCTGTCATTTAAGACCTCAAGGATGCTGGTATGCTTGTTGAAAGTGATGGTGCTC
AATGTGTTTTCATTGAAAAGTATAAGAACTAACCTCCTCTCTTCATTGTCAAAAAGGATG
GTGGTTACGGATACGATAGTACTGATATGGCCTGCATCAAGTATAGATTAAAAACTTTGA
ACTGCAATCGTTTAGTTTACATTACTGATGTTGGATAACAACCTCATTTTGATTATATTA
TTGCTGCTGCTGAAAAAATCGGATGGCACGTTCCTCCCACTACTCGTGCTGAGCATATGG
GTTTCGGTATCGTCTAAGGTGAAGACGGTAAGCGTTTCCGTACTCGTTCCAGTGAAACTG
TCAAGCTAAAAGATTTACTTGATGAAGCTAGAGAACGTGCTTTAGAAAAAATTAAAGAAC
GTATTTCAGGTGGTGCTAAAGAAGAAGGTGCTGAAGAAGATGAAGAAGAAAAAGAAAACA
AGGGTAACTACACCAAGCTCTCAGAAGAAGAATATCTTGATGCTGCTGAAAAGATGGGTA
TGGCTGCTATTAAATATTACGATTTACGTTAAAACAGAATCAGCAACTATAACTTCTCTT
ATGATAAAATGCTTGACACTAAGGGTAATACTGCTGTCTACTTACTCTATTCCTATGCTC
GTCTTTGCTCCATCATCCGTAAGAGTGGTGTCTAGTCTGAAGAACTCTGCAAAATTATCT
AAACTTAAGGATTCAAGATTTCTCATCCCCAAGAAAGAATCATTGCTTCCCTCCTCATAA
AGTTCCCTGATGTTCTTGACTAGGTTACTGAAGATTTAGCAATTAATCGTTTAACTGACT
ATATGTATGACCTTGCTTGCAAAGTAGCTGAAGGTTATCGTAAATATCACATCAACAATT
CTGAAGATAAGCTTACTCGTATCCTTCTCATTGAAGCTACTCGTTAAGTCCTTGAAAAGT
GTTTCTACTTGGTTGGTATCAAGCCCCTTGAAAAAATCTGAAAACATTAATAAATATATT
TAATATGAAACATTCAAACAAAAATTATTTTTTTATTTTATTTTACAAACATTTCTATTC
TATAAACATCTGTCATTCATTCAATCATTAATTCACTTAGCTTACTCAAAGATAAATATT
CG
>TTHERM_00657270(protein)
MESKVLKSMMKIASQHTLNVVNNALKPYNVQSPPIKIQPVSNLVDVDYNTPTLLQVFNQN
KDKKTKQAFGLADIKAFCHKVIESFNPEDKDCAIEKLGVTDQGFLTIKLKQQVIEDEINN
ILKNGLNYPTEKKQKIAVDFSSPNIAKEMHVGHLRSTIIGESICRIFEFQNHEVLRINHL
GDWGTQFGMLITYMKRKYPNFLNEPIDLSDLTMFYKESKKLFDEDPEFKKTAQLTVVDLQ
SGDPECRKAWEILCEISRREFTEIYKRLDITLEDFGESFYNDKIPAVIQDLKDAGMLVES
DGAQCVFIEKYKNQPPLFIVKKDGGYGYDSTDMACIKYRLKTLNCNRLVYITDVGQQPHF
DYIIAAAEKIGWHVPPTTRAEHMGFGIVQGEDGKRFRTRSSETVKLKDLLDEARERALEK
IKERISGGAKEEGAEEDEEEKENKGNYTKLSEEEYLDAAEKMGMAAIKYYDLRQNRISNY
NFSYDKMLDTKGNTAVYLLYSYARLCSIIRKSGVQSEELCKIIQTQGFKISHPQERIIAS
LLIKFPDVLDQVTEDLAINRLTDYMYDLACKVAEGYRKYHINNSEDKLTRILLIEATRQV
LEKCFYLVGIKPLEKI
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
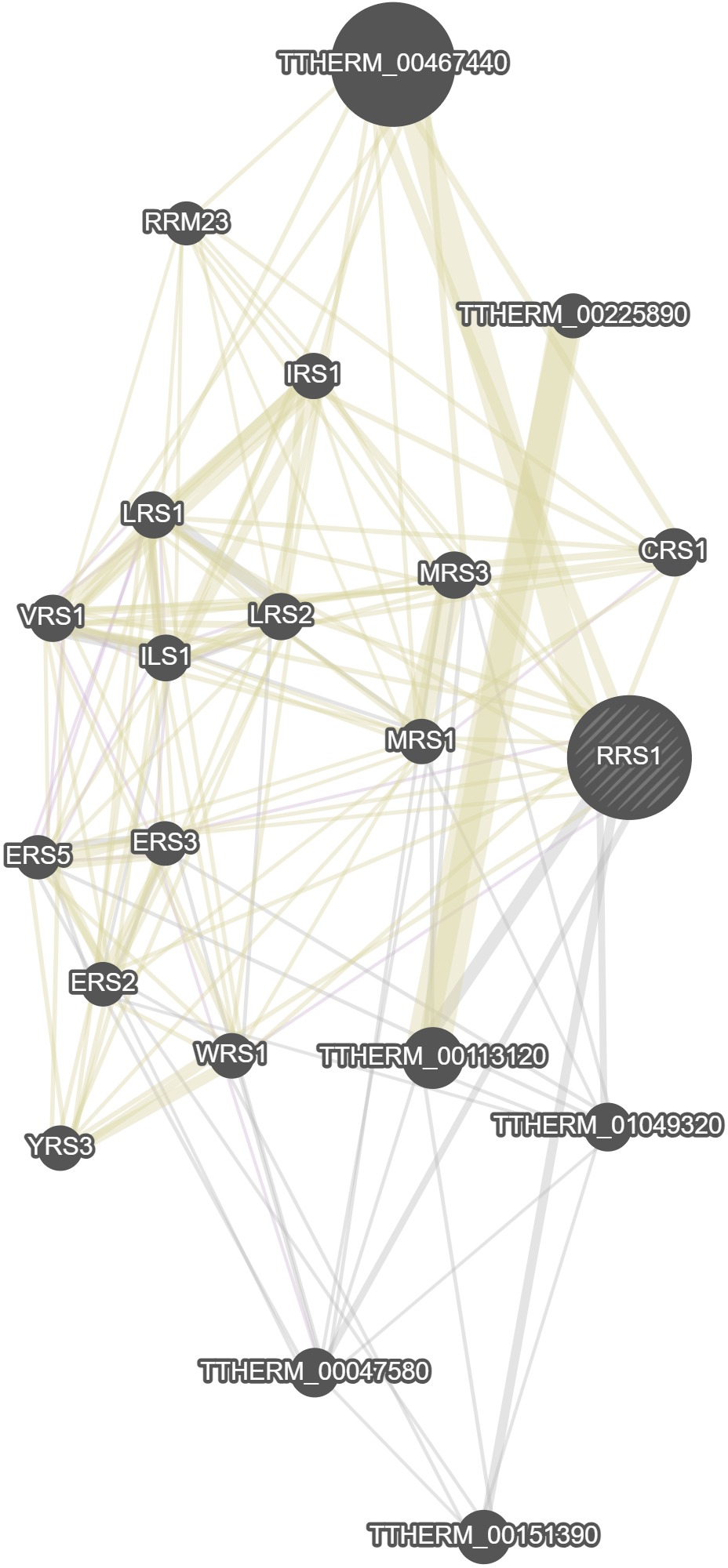
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences