Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00657460Standard Name
RRM40 (RNA recognition motif-containing protein 40)Aliases
PreTt02130 | 97.m00116 | 3724.m00024Description
RRM40 RNA recognition motif 2 in vertebrate RNA-binding protein; RRM MRD1 RNA Processing and Ribosome BiogenesisGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- cellular bud neck septin structure (IEA) | GO:0000399
- Gram-negative-bacterium-type cell wall (IEA) | GO:0009276
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- all-trans retinol 3,4-desaturase activity (IEA) | GO:0061896
- fatty acid derivative binding (IEA) | GO:1901567
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- metarhodopsin binding (IEA) | GO:0016030
- myoinhibitory hormone activity (IEA) | GO:0016085
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- open form four-way junction DNA binding (IEA) | GO:0000401
- peptide-glutamate-alpha-N-acetyltransferase activity (IEA) | GO:1990190
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- prenyltransferase activity (IEA) | GO:0004659
- prephenate dehydrogenase (NADP+) activity (IEA) | GO:0004665
- proteinase activated receptor binding (IEA) | GO:0031871
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- sodium-independent organic anion transmembrane transporter activity (IEA) | GO:0015347
- Swi5-Sfr1 complex binding (IEA) | GO:1905334
- type 1 proteinase activated receptor binding (IEA) | GO:0031872
Biological Process
- autophagy of mitochondrion (IEA) | GO:0000422
- basement membrane organization (IEA) | GO:0071711
- bronchus development (IEA) | GO:0060433
- cell migration in diencephalon (IEA) | GO:0061381
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to cocaine (IEA) | GO:0071314
- cellular response to nicotine (IEA) | GO:0071316
- coenzyme A biosynthetic process (IEA) | GO:0015937
- coenzyme A metabolic process (IEA) | GO:0015936
- epithelium development (IEA) | GO:0060429
- external genitalia morphogenesis (IEA) | GO:0035261
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- inferior colliculus development (IEA) | GO:0061379
- internal genitalia morphogenesis (IEA) | GO:0035260
- learning (IEA) | GO:0007612
- learning or memory (IEA) | GO:0007611
- long-term memory (IEA) | GO:0007616
- lung connective tissue development (IEA) | GO:0060427
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of atrichoblast fate specification (IEA) | GO:0010060
- negative regulation of autophagosome assembly (IEA) | GO:1902902
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070108
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete aging (IEA) | GO:0007568
- obsolete canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment (IEA) | GO:0061317
- obsolete canonical Wnt signaling pathway involved in positive regulation of cardiac muscle cell proliferation (IEA) | GO:0061315
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete positive regulation of mitotic G1 cell cycle arrest in response to nitrogen starvation (IEA) | GO:1903694
- obsolete positive regulation of response to DNA damage stimulus (IEA) | GO:2001022
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete senescence factor accumulation (IEA) | GO:0007579
- obsolete sodium/potassium transport (IEA) | GO:0006815
- obsolete synthesis of RNA primer involved in premeiotic DNA replication (IEA) | GO:1902980
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of double-strand break repair via homologous recombination (IEA) | GO:1905168
- positive regulation of lipoprotein particle clearance (IEA) | GO:0010986
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of protein autoubiquitination (IEA) | GO:1902499
- positive regulation of response to water deprivation (IEA) | GO:1902584
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chitin-based cuticle tanning (IEA) | GO:0007564
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of hepatocyte growth factor receptor signaling pathway (IEA) | GO:1902202
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator (IEA) | GO:1902165
- regulation of macrophage derived foam cell differentiation (IEA) | GO:0010743
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of supramolecular fiber organization (IEA) | GO:1902903
- regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903689
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- sodium-independent thromboxane transport (IEA) | GO:0071721
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- synaptic vesicle docking (IEA) | GO:0016081
- terpenoid catabolic process (IEA) | GO:0016115
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- triterpenoid catabolic process (IEA) | GO:0016105
- vesicle coating (IEA) | GO:0006901
- visual behavior (IEA) | GO:0007632
 Domains
Domains
- ( PF00076 ) RNA recognition motif
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
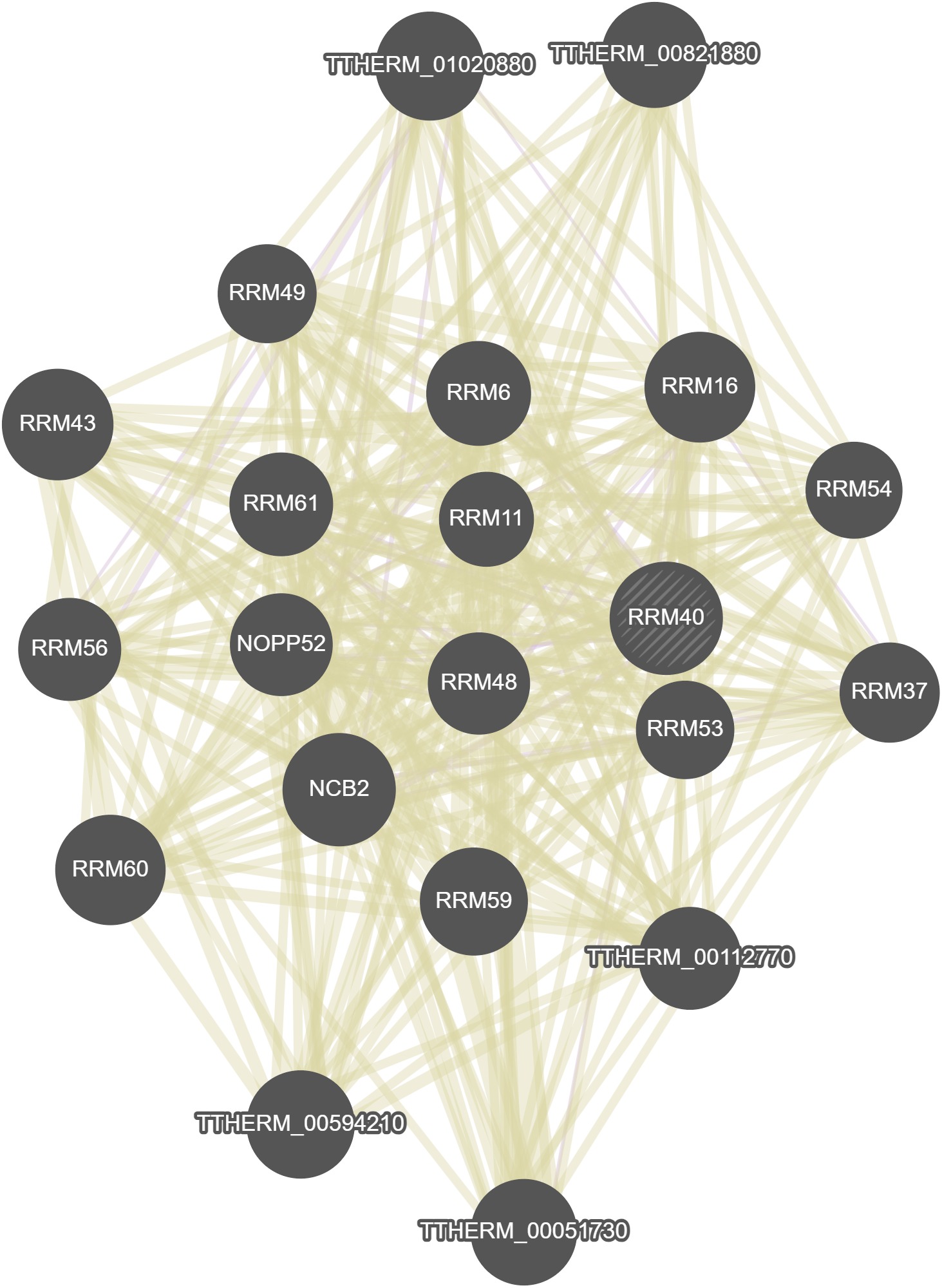
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00657460(coding)
ATGACTGACAAAAGTAAGAAAGTAGTTGAAAAACAAGAACAAGAAAACGATTAAGATAAG
AATTAAGCCCAATAAAAGAAGGGTAGAGTTATTATTAGAAATTTAATATTTGATATCAGT
GAGAAGCATCTTAAAGGTTTGTTGGGTAAGTATGGTGAAATTATTGAAATTAATATTCCA
GTCAACCCTTCAACAAACAAATCTAAGGGTTTTGCCTTTGTCTAATTTGCTAATAAAAAC
TGTGCATTGAAAGCTATAAATGAGTTAAATGGTACTTAATGGAAGGGTAGAAATATCGTT
TTAGATTTAGCTGTTTCCAAAGAAGAATTCAAAAACCATCAATAGCCATAAAATTAACAA
GATAAATAGTAATCTAAACAAAAAGCTAAAAAATCTGAATAAATTGAAGAGGAAGAAGGA
GATGACGATGAAAAAAAAATGGATGAAGAAGAAGAAAGTGAAGATGATGAAGATCAAGAA
GACGATGATGAAGAGGATGAAGACGAAGATGATGATGAAGAAGAGGAAGACGAAGATGAT
GATAAAAATAATTAAAATAAAAAGGGAAAGAATTAATAAGGATAAAAGAAATTATTTGAA
AATGAAGATTTATCCAAAACATGTTTTGTTAGAAATTTATCTTATGATTCTACTGATAAG
GATTTAAGAACCTTCTGCCAAAACTTTGGTAAAGTAGAAATGGCAAAGGTATGTATGAAT
CATGAAACAGGAAGCAACAAAGGTACTGGTTTTGTAAAATTCTAAGATGCATAAGTAGCT
CTTAAACTTATTGCAAAGAGTAATGAACTCAGTGAAAACTGTCTTTAACAAAAAAATAAA
AAAAATGCCAGCAACATTCATGATATATTTTCAGGAATAAGCGAACTTGATCTTGAAATG
AATGGAAGAAGAATAATTTTCTTGGAAGCTAAGAAAAGAGAAGAAGTTTCTAAAGTTATG
GAAGAGAAGCTTGAAAGAAATTCAATCACTTTAGATAAAGTTAAAACTTTAGATTAATTA
GAAAAGTTTGACAGATAAGGAAAGAGAAACGTTCATTTAGCCAAGCTAGGTATTTTCGAA
GAAGAAAAAAGCATGACTAATGAAGAAAAGGAAAAGAGACGTATTCACTTATAAGAAAAA
TTGAATAAGCTAAAAAACCCTAACAACTATGTATCCTCAACAAGAATTGCCCTTATGAAC
ATACCAAAAGACGAATTCTAAGAAGAAGATATTAGACTTTATGCTCAAGCAGCACTTATA
AAAGAACATGGTCAAGAATATTTTAAAAAATAAAATCCAATTAAATAGGTCTTATTGCTT
AGAGATAAGAATAAACTTGATAAGGATGGAGTACCTAAAAGCAGAGGTATTGGATTCATA
GAATTTAATACACATGAATAAGCACTTTCATTCTTGGAATATTGTGTAAATGATAAGCAA
AACTTTGAAAAGAAATACAAGAAAACACCTATTATTGAGTTTTCAATTGAAGATGCTAGA
AAAATGGCAAAAAAGAATAAATTATTAGAAAAAGTTAAATAAAAATAAGCAGAAAAGAAG
AAAGAAAAGAAAGAATTAACTGAAGAGGAATTAGAATCAAAGAAAAAAATTCTTAAAAAT
ACGAACGAAGCAGCAAAAATAAAGATTTAAAATATTCTTGATTAATCTAACTGCAGTGAC
ATGTCTCTTTATGAGGAAGGCAAGAAATTGTTGAAGGACATAAAGAGTAGAGGTATTAAA
TAAAGAATGGCCAAGCTACTAAGAAAGAAGTTCAATATTCAAAATGAAGATAATCAAAAG
TCTAATAAATTAGTAGAAGCTATTTCCAAACAAGAGGAAAAGATCAAAAATAAGAATTCT
AAGGAAGAAAAGAGAAAGCAATTATTGAAGTAGAAAGTATAAGAAGAAAAGAAGTAGAAA
GTAGCTGAATAAAAGAAGAAGCAAAAGGAAATCGATGATGAGCTTGATGTTGATGCATCT
GAACTTATTAAGGCTGTCTCTAAAGAATTAAAGAAAAATAAAATAAGAAGATAAAACTTG
GAAGACTACGATGAATTAGATGATGTAGTTGAAAAGCATATGTAAAAGAAAGTTAAAAAA
ATAAAGCAAGAAAGTAGTGCATTGTGA>TTHERM_00657460(gene)
AATAATAAATATTTGATAAAAAATAAAAGATTTCAATTAAATAGCAAGAGCAATAAAATT
TTAACAAAAATGACTGACAAAAGTAAGAAAGTAGTTGAAAAACAAGAACAAGAAAACGAT
TAAGATAAGAATTAAGCCCAATAAAAGAAGGGTAGAGTTATTATTAGAAATTTAATATTT
GATATCAGTGAGAAGCATCTTAAAGGTTTGTTGGGTAAGTATGGTGAAATTATTGAAATT
AATATTCCAGTCAACCCTTCAACAAACAAATCTAAGGGTTTTGCCTTTGTCTAATTTGCT
AATAAAAACTGTGCATTGAAAGCTATAAATGAGTTAAATGGTACTTAATGGAAGGGTAGA
AATATCGTTTTAGATTTAGCTGTTTCCAAAGAAGAATTCAAAAACCATCAATAGCCATAA
AATTAACAAGATAAATAGTAATCTAAACAAAAAGCTAAAAAATCTGAATAAATTGAAGAG
GAAGAAGGAGATGACGATGAAAAAAAAATGGATGAAGAAGAAGAAAGTGAAGATGATGAA
GATCAAGAAGACGATGATGAAGAGGATGAAGACGAAGATGATGATGAAGAAGAGGAAGAC
GAAGATGATGATAAAAATAATTAAAATAAAAAGGGAAAGAATTAATAAGGATAAAAGAAA
TTATTTGAAAATGAAGATTTATCCAAAACATGTTTTGTTAGAAATTTATCTTATGATTCT
ACTGATAAGGATTTAAGAACCTTCTGCCAAAACTTTGGTAAAGTAGAAATGGCAAAGGTA
TGTATGAATCATGAAACAGGAAGCAACAAAGGTACTGGTTTTGTAAAATTCTAAGATGCA
TAAGTAGCTCTTAAACTTATTGCAAAGAGTAATGAACTCAGTGAAAACTGTCTTTAACAA
AAAAATAAAAAAAATGCCAGCAACATTCATGATATATTTTCAGGAATAAGCGAACTTGAT
CTTGAAATGAATGGAAGAAGAATAATTTTCTTGGAAGCTAAGAAAAGAGAAGAAGTTTCT
AAAGTTATGGAAGAGAAGCTTGAAAGAAATTCAATCACTTTAGATAAAGTTAAAACTTTA
GATTAATTAGAAAAGTTTGACAGATAAGGAAAGAGAAACGTTCATTTAGCCAAGCTAGGT
ATTTTCGAAGAAGAAAAAAGCATGACTAATGAAGAAAAGGAAAAGAGACGTATTCACTTA
TAAGAAAAATTGAATAAGCTAAAAAACCCTAACAACTATGTATCCTCAACAAGAATTGCC
CTTATGAACATACCAAAAGACGAATTCTAAGAAGAAGATATTAGACTTTATGCTCAAGCA
GCACTTATAAAAGAACATGGTCAAGAATATTTTAAAAAATAAAATCCAATTAAATAGGTC
TTATTGCTTAGAGATAAGAATAAACTTGATAAGGATGGAGTACCTAAAAGCAGAGGTATT
GGATTCATAGAATTTAATACACATGAATAAGCACTTTCATTCTTGGAATATTGTGTAAAT
GATAAGCAAAACTTTGAAAAGAAATACAAGAAAACACCTATTATTGAGTTTTCAATTGAA
GATGCTAGAAAAATGGCAAAAAAGAATAAATTATTAGAAAAAGTTAAATAAAAATAAGCA
GAAAAGAAGAAAGAAAAGAAAGAATTAACTGAAGAGGAATTAGAATCAAAGAAAAAAATT
CTTAAAAATACGAACGAAGCAGCAAAAATAAAGATTTAAAATATTCTTGATTAATCTAAC
TGCAGTGACATGTCTCTTTATGAGGAAGGCAAGAAATTGTTGAAGGACATAAAGAGTAGA
GGTATTAAATAAAGAATGGCCAAGCTACTAAGAAAGAAGTTCAATATTCAAAATGAAGAT
AATCAAAAGTCTAATAAATTAGTAGAAGCTATTTCCAAACAAGAGGAAAAGATCAAAAAT
AAGAATTCTAAGGAAGAAAAGAGAAAGCAATTATTGAAGTAGAAAGTATAAGAAGAAAAG
AAGTAGAAAGTAGCTGAATAAAAGAAGAAGCAAAAGGAAATCGATGATGAGCTTGATGTT
GATGCATCTGAACTTATTAAGGCTGTCTCTAAAGAATTAAAGAAAAATAAAATAAGAAGA
TAAAACTTGGAAGACTACGATGAATTAGATGATGTAGTTGAAAAGCATATGTAAAAGAAA
GTTAAAAAAATAAAGCAAGAAAGTAGTGCATTGTGAAATCATTTTAATCAAACAATAATT
ACAAATCATTAAAAATATTGAGTTAATATTAGAATTTTAGGTGCTTATGATATGCATCTT
GAAATTTAAGTAAAGTTGTAATTAAACGAAAGTAGGTTTAGGGATTAAACTTTATTACTT
TTTATAAAATTTTAATTATAGTAAAATTTGATATGTCAATTTCTTCAATATATTGTATTC
TAGATATTTTATGGTTTATATCCTTAATTCTTTTTTAAGAACCTTTTATTATCAGCTTTG
ATAGAAAAGGTATATACTCTTTGCAAATAA>TTHERM_00657460(protein)
MTDKSKKVVEKQEQENDQDKNQAQQKKGRVIIRNLIFDISEKHLKGLLGKYGEIIEINIP
VNPSTNKSKGFAFVQFANKNCALKAINELNGTQWKGRNIVLDLAVSKEEFKNHQQPQNQQ
DKQQSKQKAKKSEQIEEEEGDDDEKKMDEEEESEDDEDQEDDDEEDEDEDDDEEEEDEDD
DKNNQNKKGKNQQGQKKLFENEDLSKTCFVRNLSYDSTDKDLRTFCQNFGKVEMAKVCMN
HETGSNKGTGFVKFQDAQVALKLIAKSNELSENCLQQKNKKNASNIHDIFSGISELDLEM
NGRRIIFLEAKKREEVSKVMEEKLERNSITLDKVKTLDQLEKFDRQGKRNVHLAKLGIFE
EEKSMTNEEKEKRRIHLQEKLNKLKNPNNYVSSTRIALMNIPKDEFQEEDIRLYAQAALI
KEHGQEYFKKQNPIKQVLLLRDKNKLDKDGVPKSRGIGFIEFNTHEQALSFLEYCVNDKQ
NFEKKYKKTPIIEFSIEDARKMAKKNKLLEKVKQKQAEKKKEKKELTEEELESKKKILKN
TNEAAKIKIQNILDQSNCSDMSLYEEGKKLLKDIKSRGIKQRMAKLLRKKFNIQNEDNQK
SNKLVEAISKQEEKIKNKNSKEEKRKQLLKQKVQEEKKQKVAEQKKKQKEIDDELDVDAS
ELIKAVSKELKKNKIRRQNLEDYDELDDVVEKHMQKKVKKIKQESSAL