Gene Model Identifier
TTHERM_00664030
Standard Name
DESI1
(DeSumoylating Isopeptidase 1)
Aliases
PreTt23850 | 98.m00193 | 3736.m00091 | LC3A
Description
DESI1 thioredoxin domain protein; SUMO-specific protease; deSUMOylase; removes Smt3p from protein substrates; Desi1p is distinct from the Sentrin-specific proteases (SENP) family of deSUMOylases; PPPDE peptidase domain
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00664030(coding)
ATGGCTGAACAAAAGCATAGGGTAGTAGTTAATTTGTATGATTTATCTGGAGGTATGGCT
AGAGTTTTCTCTCCCATGTTCCTTTAGAAACAAATTGATGGAATTTGGCACACTGGTTGT
GTTGTTTACGGTAAGGAATTTTATTTTGGTGGAGGCATCTGCTCTGGCTTACCAAAATAG
ACACCTTACGGAACACCAGTTTAGTAAATAGATGTTGGAGAGACTGAAGTTCCTGAAGAA
GTCTTTACAGAGTTTTTGAGAGATATTTCAGATAGATTTACTATGGATAAATATGACTTA
TTTAAAAATAATTGCAACAACTTTACTGATGAATGTACTCATTTCCTAACAGGATAACAT
ATTCCTGAGTATATAACAGGTTTACCTGATGAAGTATTAAATACTCCTATGGGTCAAATG
ATAAAACCTCTTGTTGATCAAATGCAATAATCCGTTGTAAATGCAAGTGGTTAATCAGCA
CTATTCCCAGCTTAGTTTGAAGGTGTTAATAATCCTCAATAAATGTTTGGAAATAATTAA
CAATAATAATAATAATAGCAATAGGCACCTATTTTCCCTAATGGTATAAATAATACAGAT
ATCCTCAATAAATTTACTTAGTTGAATAATAACTAACCAGTTGATATGAAGAAAGTTGAA
GAAGCAACTGATTTAATGGATTACTTTACTAAGGTAGAGGGACAAAAAGCAGTGATTATC
GACTTTAACGCTGATTGGTGTGGACCTTGTTAAGTAATCAAGCCAATATTTGCTTAATAT
AGTGAAGAATACCCAAATATCAAATTTATTTCTGTAAATACAGAAAAAAACAAGGAGGTT
GCCCAATAATTTGGCATTTAATCTCTCCCCACCTTCATCACCCTTCATGAAGGTGAAATA
TCTGCAACTTGGAAAGGAGCTAATCAAGTAAACTTAAAAAATAACCTTGATAAATTAGCA
GAAAAGCTTAAATGA
>TTHERM_00664030(gene)
TTTCAAAATTTTGTTCTTCAGTAATTATAAATATTAATAAATAATAGGAAGGAGGGAGAT
GGATTTGCTAGTATAAATAAATAAATAACTATATTAATAAAACTAAAAGTTTTTTTATTT
AAAGGAACACAATAGATTTAGAAGAGATGTAAAAGGAAAGAATCGAATTACGATATCAAA
AGAATAGATTATTTAGATAGACTGACTCTAAATTGAGTATTTGAAAAGCATCAACGATAA
ACATAATTTTTATTTAAATTGTTAAGAATTAGTAAAAAAAGAGAAAAGGTTTTTAAATTT
TCTTAAATTTATTCAGATTTCACCAAAATATTAAAAAATAAAAGCAATAAAATAAAATAA
GTTTTTAAATGGCTGAACAAAAGCATAGGGTAGTAGTTAATTTGTATGATTTATCTGGAG
GTATGGCTAGAGTTTTCTCTCCCATGTTCCTTTAGAAACAAATTGATGGAATTTGGCACA
CTGGTTGTGTTGTTTACGGTAAGGAATTTTATTTTGGTGGAGGCATCTGCTCTGGCTTAC
CAAAATAGACACCTTACGGAACACCAGTTTAGTAAATAGATGTTGGAGAGACTGAAGTTC
CTGAAGAAGTCTTTACAGAGTTTTTGAGAGATATTTCAGATAGATTTACTATGGATAAAT
ATGACTTATTTAAAAATAATTGCAACAACTTTACTGATGAATGTACTCATTTCCTAACAG
GATAACATATTCCTGAGTATATAACAGGTTTACCTGATGAAGTATTAAATACTCCTATGG
GTCAAATGATAAAACCTCTTGTTGATCAAATGCAATAATCCGTTGTAAATGCAAGTGGTT
AATCAGCACTATTCCCAGCTTAGTTTGAAGGTGTTAATAATCCTCAATAAATGTTTGGAA
ATAATTAACAATAATAATAATAATAGCAATAGGCACCTATTTTCCCTAATGGTATAAATA
ATACAGATATCCTCAATAAATTTACTTAGTTGAATAATAACTAACCAGTTGATATGAAGA
AAGTTGAAGAAGCAACTGATTTAATGGATTACTTTACTAAGGTAGAGGGACAAAAAGCAG
TGATTATCGACTTTAACGCTGATTGGTGTGGACCTTGTTAAGTAATCAAGCCAATATTTG
CTTAATATAGTGAAGAATACCCAAATATCAAATTTATTTCTGTAAATACAGAAAAAAACA
AGGAGGTTGCCCAATAATTTGGCATTTAATCTCTCCCCACCTTCATCACCCTTCATGAAG
GTGAAATATCTGCAACTTGGAAAGGAGCTAATCAAGTAAACTTAAAAAATAACCTTGATA
AATTAGCAGAAAAGCTTAAATGAAAAATGATTAAAGTATTTGATTCAGTATATGAAAATG
TTTTATAAAAATTTATGTTTTACATTTAAATGTATAATATTTATAATAAATAAAAT
>TTHERM_00664030(protein)
MAEQKHRVVVNLYDLSGGMARVFSPMFLQKQIDGIWHTGCVVYGKEFYFGGGICSGLPKQ
TPYGTPVQQIDVGETEVPEEVFTEFLRDISDRFTMDKYDLFKNNCNNFTDECTHFLTGQH
IPEYITGLPDEVLNTPMGQMIKPLVDQMQQSVVNASGQSALFPAQFEGVNNPQQMFGNNQ
QQQQQQQQAPIFPNGINNTDILNKFTQLNNNQPVDMKKVEEATDLMDYFTKVEGQKAVII
DFNADWCGPCQVIKPIFAQYSEEYPNIKFISVNTEKNKEVAQQFGIQSLPTFITLHEGEI
SATWKGANQVNLKNNLDKLAEKLK
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
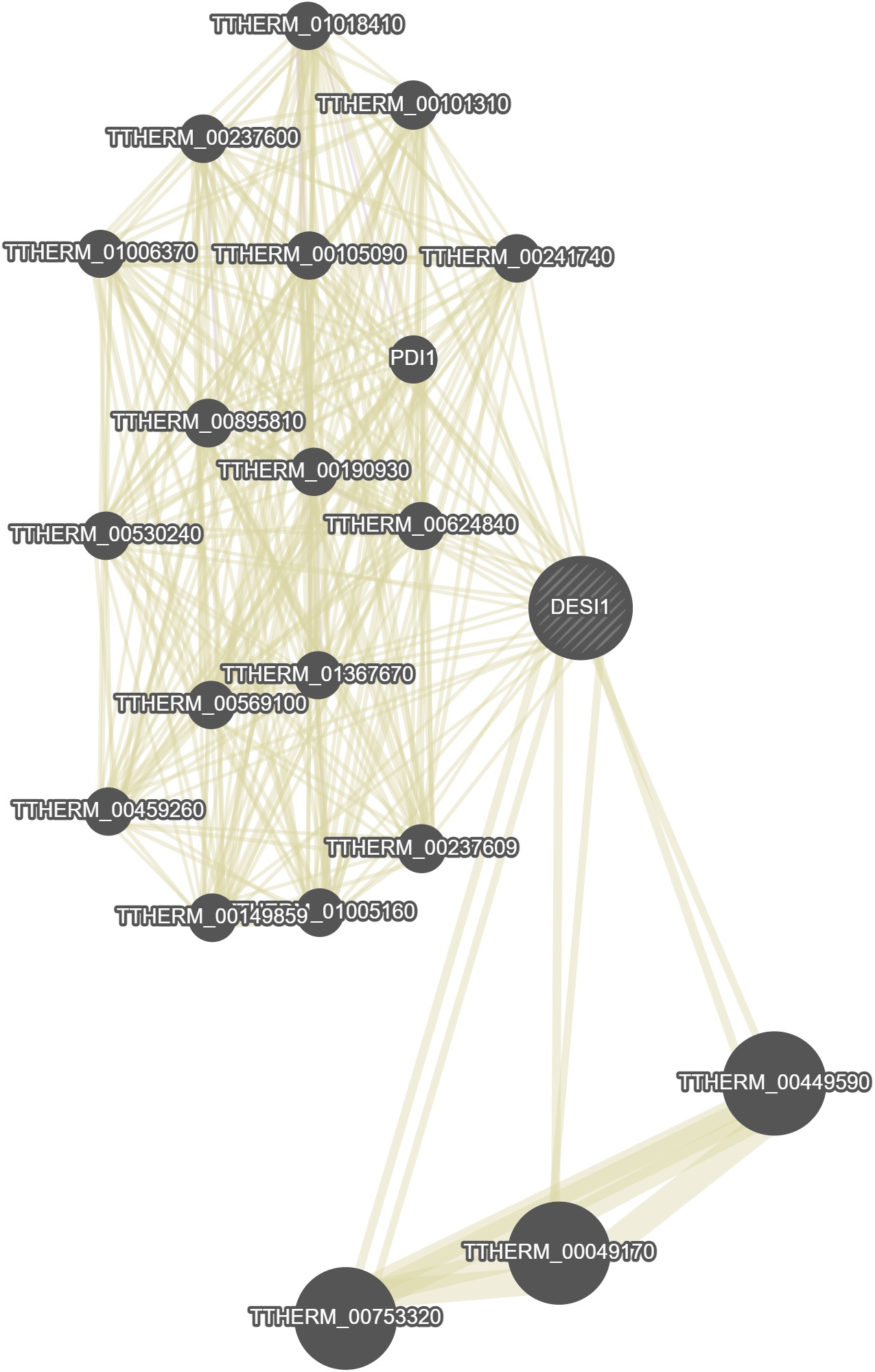
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences