Gene Model Identifier
TTHERM_00683070
Standard Name
RRM43
(RNA recognition motif-containing protein 43)
Aliases
PreTt16144 | 103.m00135 | 3734.m00047
Description
RRM43 RNA-binding domain protein; TatSF1-like
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
>TTHERM_00683070(coding)
ATGGAGGAATTTATTTGGTATTATGCAGATAGTTAAGCTCTGTAAGAGAGAACACCTATA
GGACCTGTTAGCATTAGAGATTTAGATGTTTTGTATAGGACTTCAGCAATAAATTCTGCA
ACTTATGTTTGGAAAGAAGGTATGCCTGAATGGGTTTAACTTTTTAAAGTATAGGAATTG
AAGGAGGCTATTCTAGACGAGCAATAAGATATTAAGGTAATATAAGAACTTTATGAAAAG
TAGTAGCAGGAACTAGCTCTAGCTAATTAAAAGGGTAAAAAGTAATAGAAAATACAAGAT
AAACTTGACAAAAAATAAGCTTAGATTGATCAAAAGTTGCTTGAGGAAGCTAAATATAAT
TAATCAATGAATAGTGGCAAGGAGGAAGATATAAGTCAAGATTAGGTAAAATAAAACTTG
GAATTGTTATAGCAAATAGATGAAGAAGAAGTGTTAGATGAAGATGAAGAAGAAGAAAGG
GAGTATTTAAAGCTGTTATAAGAAAATCCAGAAGAAGCGATTTAAAGAATGGCCAATAAC
ACTTTAAACAGCTATTTTTACTATTCTGAAGATGATAAAAAGTGGAATATTTGTAGAGAA
GATGAAAATAATGAGAGAGTTTGGATCTAATAAGATGAAGAACCTAAAGAAGAAATGGAT
AAAATGAGAAATGAATTGATAGAAAAGCTTAGAAAATAATTTATAGGTGAAGAAAACAAA
GATATTGAGACATTGAAAAGCAATTTGAAAGATCAGCTATAAATGTAAAATGATATTGAA
GATAATAACGCTGATCCTAAAGATAAAGAAATTATGAAAGGTATGACAGCAGAGCAAATT
AAAAAGATAAAACGTAACAAAAAGAAAGCCAAAAGATAAGCTGAAAAAAAGAAATAGAAA
TGGTATTAATCTAGGGTTAATACATATATATATGTAAAGGGATTGCCTCTTTCTATTACA
GAAGAAAAACTCGATGAATTCTTTTCCAGAGCAGGAGTAATTAGAAAGGACCCAATCACC
AATAAAAAAAAGATCAAAATTTACTCGGATGAAAACGGTCTCCCTAAAGGTGATGCAGTT
ATCAGCTTTTAGATGATGGAATCTGTAGAAATTGCAATTACTATGCTTGATGAGAGAGAA
ATAGAACCAGGCCATATTATTAGGGTCGAAAGAGCTAATTTTGAATAGCATGGAGATACT
TATAAGAAAAGAGAAGGTGTTGTTATTAACAAAGAAAAAGCTTTGGATAAAATTTAACTT
GCCTAAATGAAAGCTGCTCAAAGATAGGCTCTGGGTTGGGAAGATGAAGACCAAATTGAC
ACAGGACTTAAAATTGTCATATTAAAAAATGTATTTACTCTTAAGGATATAGAAGAAGAT
CCAAATTTCTTGGAAGAACTCAGAGAAGAAATGGCTAAAGAGATAGAGTCAACTTGTGGA
CCAATTTAGAGATTAAAAATATTTGAAGAAAATCCAGAAGGAGTTATTGAAATTAAATTT
AAAAATTCTGCTGATGCCAAGGTTTGCATTCAAAAAATGGATGGTAGATATTTTGATGAG
AGAGAACTTGAGTGCTTTTTCTGGGATGGTAAGGTAGATTACAAAAGGTCTTAAAAACAA
ATGGAAGATGACGAATAAAGACTTGAAGAATTTGGTAAATGGATAGAAGGAGGTGATAAT
AATAATTCAGAAAATGGTTAAAAATAAATTAGTTGA
>TTHERM_00683070(gene)
TTATATTGTATATCAAATAAATAAAGATACATTAAATAAAAAAAAATATTTATATAGTAA
ATTCAGTTAATATTAAGTTAATAAAGTTTTATGGAGGAATTTATTTGGTATTATGCAGAT
AGTTAAGCTCTGTAAGAGAGAACACCTATAGGACCTGTTAGCATTAGAGATTTAGATGTT
TTGTATAGGACTTCAGCAATAAATTCTGCAACTTATGTTTGGAAAGAAGGTATGCCTGAA
TGGGTTTAACTTTTTAAAGTATAGGAATTGAAGGAGGCTATTCTAGACGAGCAATAAGAT
ATTAAGGTAATATAAGAACTTTATGAAAAGTAGTAGCAGGAACTAGCTCTAGCTAATTAA
AAGGGTAAAAAGTAATAGAAAATACAAGATAAACTTGACAAAAAATAAGCTTAGATTGAT
CAAAAGTTGCTTGAGGAAGCTAAATATAATTAATCAATGAATAGTGGCAAGGAGGAAGAT
ATAAGTCAAGATTAGGTAAAATAAAACTTGGAATTGTTATAGCAAATAGATGAAGAAGAA
GTGTTAGATGAAGATGAAGAAGAAGAAAGGGAGTATTTAAAGCTGTTATAAGAAAATCCA
GAAGAAGCGATTTAAAGAATGGCCAATAACACTTTAAACAGCTATTTTTACTATTCTGAA
GATGATAAAAAGTGGAATATTTGTAGAGAAGATGAAAATAATGAGAGAGTTTGGATCTAA
TAAGATGAAGAACCTAAAGAAGAAATGGATAAAATGAGAAATGAATTGATAGAAAAGCTT
AGAAAATAATTTATAGGTGAAGAAAACAAAGATATTGAGACATTGAAAAGCAATTTGAAA
GATCAGCTATAAATGTAAAATGATATTGAAGATAATAACGCTGATCCTAAAGATAAAGAA
ATTATGAAAGGTATGACAGCAGAGCAAATTAAAAAGATAAAACGTAACAAAAAGAAAGCC
AAAAGATAAGCTGAAAAAAAGAAATAGAAATGGTATTAATCTAGGGTTAATACATATATA
TATGTAAAGGGATTGCCTCTTTCTATTACAGAAGAAAAACTCGATGAATTCTTTTCCAGA
GCAGGAGTAATTAGAAAGGACCCAATCACCAATAAAAAAAAGATCAAAATTTACTCGGAT
GAAAACGGTCTCCCTAAAGGTGATGCAGTTATCAGCTTTTAGATGATGGAATCTGTAGAA
ATTGCAATTACTATGCTTGATGAGAGAGAAATAGAACCAGGCCATATTATTAGGGTCGAA
AGAGCTAATTTTGAATAGCATGGAGATACTTATAAGAAAAGAGAAGGTGTTGTTATTAAC
AAAGAAAAAGCTTTGGATAAAATTTAACTTGCCTAAATGAAAGCTGCTCAAAGATAGGCT
CTGGGTTGGGAAGATGAAGACCAAATTGACACAGGACTTAAAATTGTCATATTAAAAAAT
GTATTTACTCTTAAGGATATAGAAGAAGATCCAAATTTCTTGGAAGAACTCAGAGAAGAA
ATGGCTAAAGAGATAGAGTCAACTTGTGGACCAATTTAGAGATTAAAAATATTTGAAGAA
AATCCAGAAGGAGTTATTGAAATTAAATTTAAAAATTCTGCTGATGCCAAGGTTTGCATT
CAAAAAATGGATGGTAGATATTTTGATGAGAGAGAACTTGAGTGCTTTTTCTGGGATGGT
AAGGTAGATTACAAAAGGTCTTAAAAACAAATGGAAGATGACGAATAAAGACTTGAAGAA
TTTGGTAAATGGATAGAAGGAGGTGATAATAATAATTCAGAAAATGGTTAAAAATAAATT
AGTTGATTTAAATTTAAGAAGTTATTAGCTTGAAAATCTTTTTTTTTTTTGAAATAAGGA
GTTTTATATGTGAAAATTTTTAGTTGAAATTTGATTTATTTGAAAATCTTAATTACTTTA
TTATTGAGATATATATGTATGAATG
>TTHERM_00683070(protein)
MEEFIWYYADSQALQERTPIGPVSIRDLDVLYRTSAINSATYVWKEGMPEWVQLFKVQEL
KEAILDEQQDIKVIQELYEKQQQELALANQKGKKQQKIQDKLDKKQAQIDQKLLEEAKYN
QSMNSGKEEDISQDQVKQNLELLQQIDEEEVLDEDEEEEREYLKLLQENPEEAIQRMANN
TLNSYFYYSEDDKKWNICREDENNERVWIQQDEEPKEEMDKMRNELIEKLRKQFIGEENK
DIETLKSNLKDQLQMQNDIEDNNADPKDKEIMKGMTAEQIKKIKRNKKKAKRQAEKKKQK
WYQSRVNTYIYVKGLPLSITEEKLDEFFSRAGVIRKDPITNKKKIKIYSDENGLPKGDAV
ISFQMMESVEIAITMLDEREIEPGHIIRVERANFEQHGDTYKKREGVVINKEKALDKIQL
AQMKAAQRQALGWEDEDQIDTGLKIVILKNVFTLKDIEEDPNFLEELREEMAKEIESTCG
PIQRLKIFEENPEGVIEIKFKNSADAKVCIQKMDGRYFDERELECFFWDGKVDYKRSQKQ
MEDDEQRLEEFGKWIEGGDNNNSENGQKQIS
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
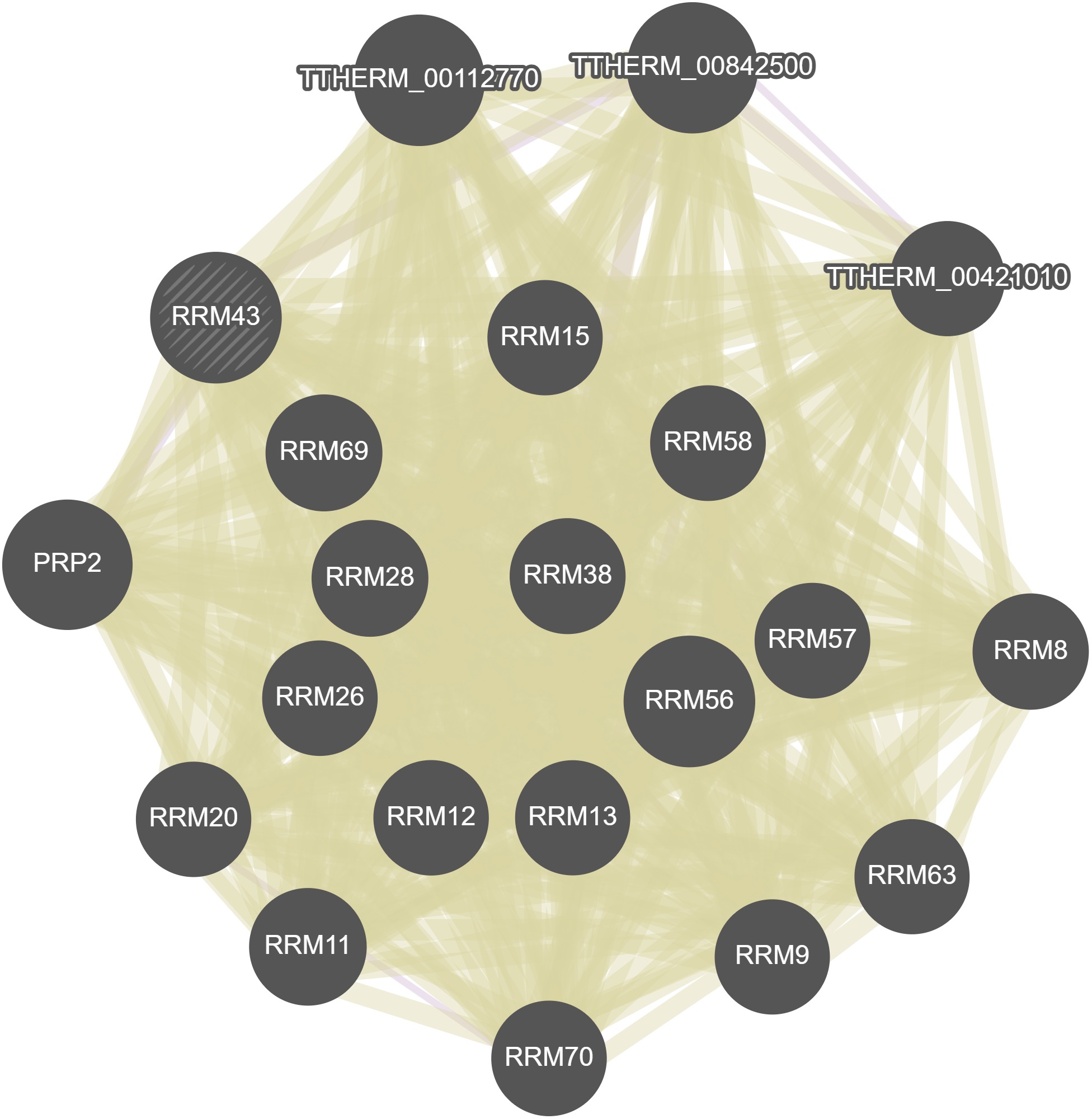
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences