Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00691200Standard Name
TFIISAliases
PreTt02958 | 106.m00098 | 3674.m00011Description
TFIIS transcription factor S-II; transcription factor S-II; Interacts with transcription regulator Mediator complex in Tetrahymena; Zinc finger, TFIIS-typeGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- catalase complex (IEA) | GO:0062151
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- phagophore (IEA) | GO:0061908
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- all-trans retinol 3,4-desaturase activity (IEA) | GO:0061896
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- metarhodopsin binding (IEA) | GO:0016030
- MHC class II receptor activity (IEA) | GO:0032395
- microtubule severing ATPase activity (IEA) | GO:0008568
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- proteinase activated receptor binding (IEA) | GO:0031871
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- Swi5-Sfr1 complex binding (IEA) | GO:1905334
Biological Process
- basement membrane organization (IEA) | GO:0071711
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- coenzyme A biosynthetic process (IEA) | GO:0015937
- coenzyme A metabolic process (IEA) | GO:0015936
- endoderm development (IEA) | GO:0007492
- epiboly involved in gastrulation with mouth forming second (IEA) | GO:0055113
- epithelium development (IEA) | GO:0060429
- evasion or tolerance by organism of reactive oxygen species produced by other organism involved in symbiotic interaction (IEA) | GO:0052385
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- high-affinity zinc II ion transport (IEA) | GO:0006830
- homogentisate metabolic process (IEA) | GO:1901999
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- induction by organism of defense-related reactive oxygen species production in other organism involved in symbiotic interaction (IEA) | GO:0052264
- internal genitalia morphogenesis (IEA) | GO:0035260
- lung connective tissue development (IEA) | GO:0060427
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- mammary gland epithelium development (IEA) | GO:0061180
- mesodermal cell fate determination (IEA) | GO:0007500
- midgut development (IEA) | GO:0007494
- negative regulation by organism of jasmonic acid-mediated defense response of other organism involved in symbiotic interaction (IEA) | GO:0052266
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of antimicrobial peptide secretion (IEA) | GO:0002795
- negative regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903885
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of integrin-mediated signaling pathway (IEA) | GO:2001045
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of protein localization to presynapse (IEA) | GO:1905385
- negative regulation of response to drug (IEA) | GO:2001024
- negative regulation of silent mating-type cassette heterochromatin formation (IEA) | GO:0061186
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- obsolete age dependent accumulation of genetic damage (IEA) | GO:0007570
- obsolete age-dependent yeast cell size increase (IEA) | GO:0007581
- obsolete aging (IEA) | GO:0007568
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete positive regulation of diuresis by angiotensin (IEA) | GO:0003102
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete ribonucleoprotein complex import into nucleus (IEA) | GO:0071167
- obsolete senescence factor accumulation (IEA) | GO:0007579
- obsolete small molecule transport (IEA) | GO:0006832
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- parturition (IEA) | GO:0007567
- phenylpropanoid biosynthetic process (IEA) | GO:0009699
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of aconitate hydratase activity (IEA) | GO:1904234
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of protein autoubiquitination (IEA) | GO:1902499
- positive regulation of response to water deprivation (IEA) | GO:1902584
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of chitin-based cuticle tanning (IEA) | GO:0007564
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of phosphorus utilization (IEA) | GO:0006795
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- snoRNA guided rRNA 2'-O-methylation (IEA) | GO:0000452
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- synaptic vesicle docking (IEA) | GO:0016081
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- triterpenoid catabolic process (IEA) | GO:0016105
- vesicle coating (IEA) | GO:0006901
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
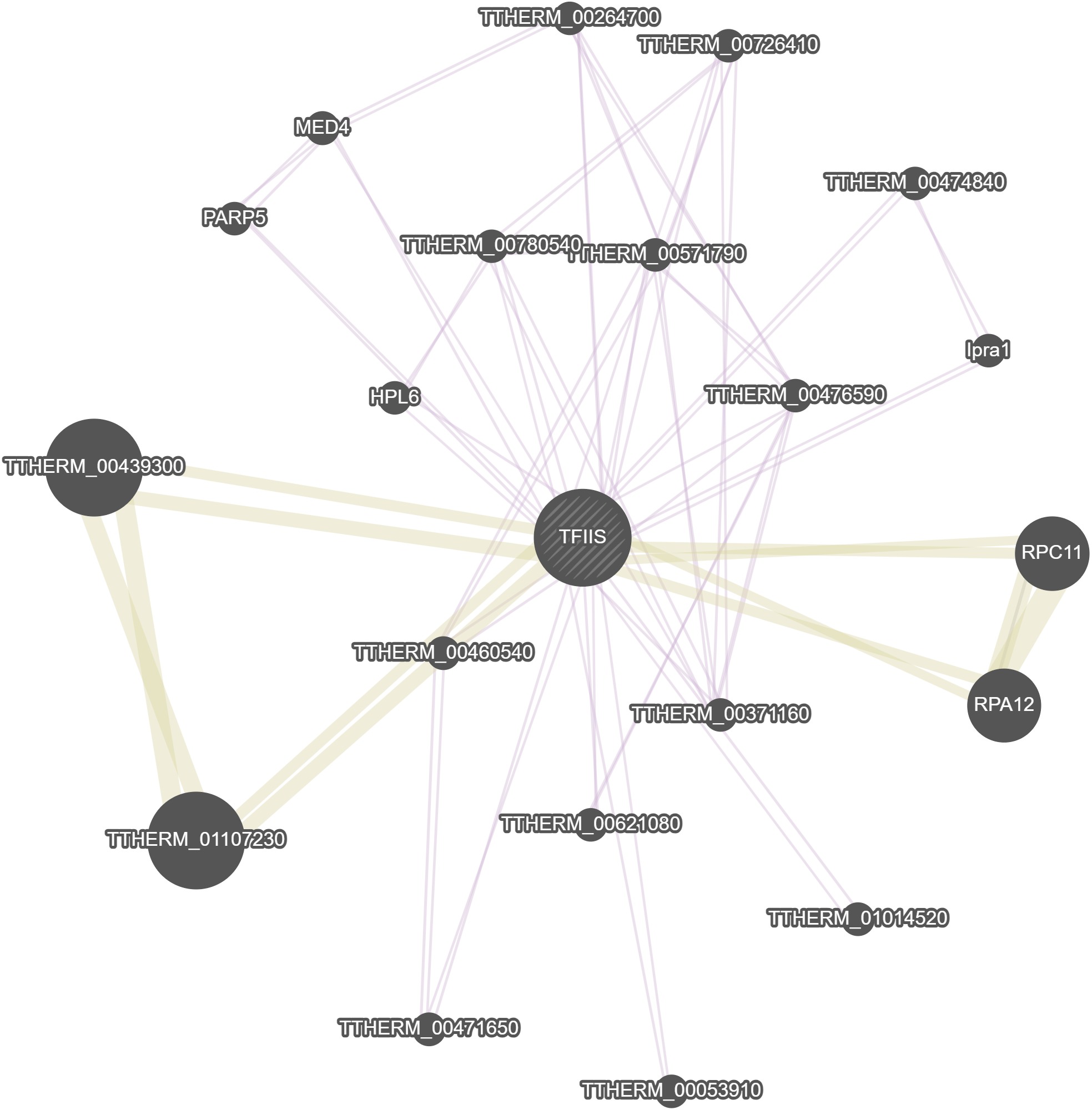
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:31280994: Garg J, Saettone A, Nabeel-Shah S, Cadorin M, Ponce M, Marquez S, Pu S, Greenblatt J, Lambert JP, Pearlman RE, Fillingham J (2019) The Med31 Conserved Component of the Divergent Mediator Complex in Tetrahymena thermophila Participates in Developmental Regulation. Current biology : CB 29(14):2371-2379.e6
 Sequences
Sequences
>TTHERM_00691200(coding)
ATGGATCAACTATACACAGAAGATGAATTGACTGAATTCAAAAACGATTTGAGAAAGTTT
AAGGAAATTGGAAATACTAAAGTTTTGGCCATCGTAAAAAGACTTTTTAATTGCCAAATC
ACAGTATAATAATTATAAAACACTAAAATTGCTTAAACTGTGTAATCTCTCTCTACTATG
TCTTGCTTCGATGAAGATGAAGAAGGACAAGAAGCTAAATAAATATGCTCTTAAATTATT
AACAAATGGAAAAAAATAGTTTAAAATGAAAAAAATAAGCAAAATCCTCAAGCCTCAAGC
TAAAAAGCACCACAATTAACTAAATCAAATTCACAAACTGCTACTTTGCCTAGAAGATAA
TCCTCTGGTTAAGGTTCTACTGAAACATTTTTAACTAATGAGGAACCCTAATACAAGACA
GAATCAAAAACTGATCTTCCTACTTATAAAAGTGATATTCGCTAAAAAGGATTAGAAGCT
ATTAACAAAATTTTTATGAAGGTTAAAGATGATATTAATGCTTCTCCTAGAGAATGGATT
GAATTAATTGTAAATCTTGAAAAGAAAATTTTCTCAAGAAATTCCTAAGTAGTTTACTAT
AAATCTTATATTAAAGAAGTCCTTGGTATGTTCTCTTCTAATGAAGTTACCACTGAGGTT
ATTTCTAAAATGTTAAATAAAGAAATTTCACTTTAGGATGTTGCTGAAGCAAAAACTAGT
GTCTTCTTGACTGATAGATAAAGAGCCTAGCAAAAAAAAGCTGTTGAAGATTAGCTTGCT
ACAAGTGATCCAGATTTTTATAACAATATGCGTAGATAGCGTTTACAAGGTCTAGAAGGT
GAATTATGTAAAGGATGTAAGAAAAAAACAGCTTTCTTAGTAAAAGAATTATAGACTCGT
TCATCAGATGAACCTATGACTCGTTTTATGGAATGCAATAGCTGTGGTAAAAGTTGGAAA
GACTGA>TTHERM_00691200(gene)
TCCTTCTATTAAATTGAATGTTTAAATAAAATTTACATTAGATTTTTCCATTTGGGGCTA
TTTATGAATACTGACATTCAATTTTGAGCTTTTAAGCGCAAAAAAATCAAAGCTAAACAT
TCTTAAAAACTTTAAAATATTCTTTAATCAAACACAAATCTTAATATAAAGGTAAAAAAT
ATAAATTTCATAGCAATTTTACTAAATATTAAAAAAGAAAGGCAGGAAAATTTAAGAATA
GATTTGAAAATAAAATGGATCAACTATACACAGAAGATGAATTGACTGAATTCAAAAACG
ATTTGAGAAAGTTTAAGGAAATTGGAAATACTAAAGTTTTGGCCATCGTAAAAAGACTTT
TTAATTGCCAAATCACAGTATAATAATTATAAAACACTAAAATTGCTTAAACTGTGTAAT
CTCTCTCTACTATGTCTTGCTTCGATGAAGATGAAGAAGGACAAGAAGCTAAATAAATAT
GCTCTTAAATTATTAACAAATGGAAAAAAATAGTTTAAAATGAAAAAAATAAGCAAAATC
CTCAAGCCTCAAGCTAAAAAGCACCACAATTAACTAAATCAAATTCACAAACTGCTACTT
TGCCTAGAAGATAATCCTCTGGTTAAGGTTCTACTGAAACATTTTTAACTAATGAGGAAC
CCTAATACAAGACAGAATCAAAAACTGATCTTCCTACTTATAAAAGTGATATTCGCTAAA
AAGGATTAGAAGCTATTAACAAAATTTTTATGAAGGTTAAAGATGATATTAATGCTTCTC
CTAGAGAATGGATTGAATTAATTGTAAATCTTGAAAAGAAAATTTTCTCAAGAAATTCCT
AAGTAGTTTACTATAAATCTTATATTAAAGAAGTCCTTGGTATGTTCTCTTCTAATGAAG
TTACCACTGAGGTTATTTCTAAAATGTTAAATAAAGAAATTTCACTTTAGGATGTTGCTG
AAGCAAAAACTAGTGTCTTCTTGACTGATAGATAAAGAGCCTAGCAAAAAAAAGCTGTTG
AAGATTAGCTTGCTACAAGTGATCCAGATTTTTATAACAATATGCGTAGATAGCGTTTAC
AAGGTCTAGAAGGTGAATTATGTAAAGGATGTAAGAAAAAAACAGCTTTCTTAGTAAAAG
AATTATAGACTCGTTCATCAGATGAACCTATGACTCGTTTTATGGAATGCAATAGCTGTG
GTAAAAGTTGGAAAGACTGAAGGAAGTAAAAGAAGGAGAATAAATAATCTTTTAAAATCA
TAGCTAAAAATTCAAATAATTTTGAATTAAAAATAGTTACAATCGTATTAAGCTATATAG
TAAAATATAATTTTGAGAAAACTGAGCAAACTAACTAACCAGATATAATTTAAATATTAT
TTAAACATACTAACTCAATCATTCATTATTCAAAGTTTTAAGTTTGGATATGAATAATTT
ATTATAGTATATATAATCATTCATAATGCTTGCATACTTCTAAAATTTTGTGAGTTTCCT
AATTTTA>TTHERM_00691200(protein)
MDQLYTEDELTEFKNDLRKFKEIGNTKVLAIVKRLFNCQITVQQLQNTKIAQTVQSLSTM
SCFDEDEEGQEAKQICSQIINKWKKIVQNEKNKQNPQASSQKAPQLTKSNSQTATLPRRQ
SSGQGSTETFLTNEEPQYKTESKTDLPTYKSDIRQKGLEAINKIFMKVKDDINASPREWI
ELIVNLEKKIFSRNSQVVYYKSYIKEVLGMFSSNEVTTEVISKMLNKEISLQDVAEAKTS
VFLTDRQRAQQKKAVEDQLATSDPDFYNNMRRQRLQGLEGELCKGCKKKTAFLVKELQTR
SSDEPMTRFMECNSCGKSWKD