 Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00700900
Standard Name
TPA2
(Tetrahymena P-type ATPase )
Aliases
PreTt14887 | 109.m00105
Description
TPA2 Na-H K antiporter P-type ATPase alpha subunit family protein; P-type ATPase; putative Na(+)/K(+) or H(+)/K(+) ATPase; constitutively expressed; expression upregulated at 37C and downregulated by starvation; Cation transport ATPase (P-type)
Genome
Browser (Macronucleus)
No Genome Browser Data Present
Genome Browser (Micronucleus)
No Genome Browser Data Present
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
Molecular Function
 Domains
Domains
- ( PF00122 ) E1-E2 ATPase
- ( PF00689 ) Cation transporting ATPase, C-terminus
- ( PF00690 ) Cation transporter/ATPase, N-terminus
- ( PF13246 ) Cation transport ATPase (P-type)
 Gene Expression Profile
Gene Expression Profile
 GeneMania
GeneMania
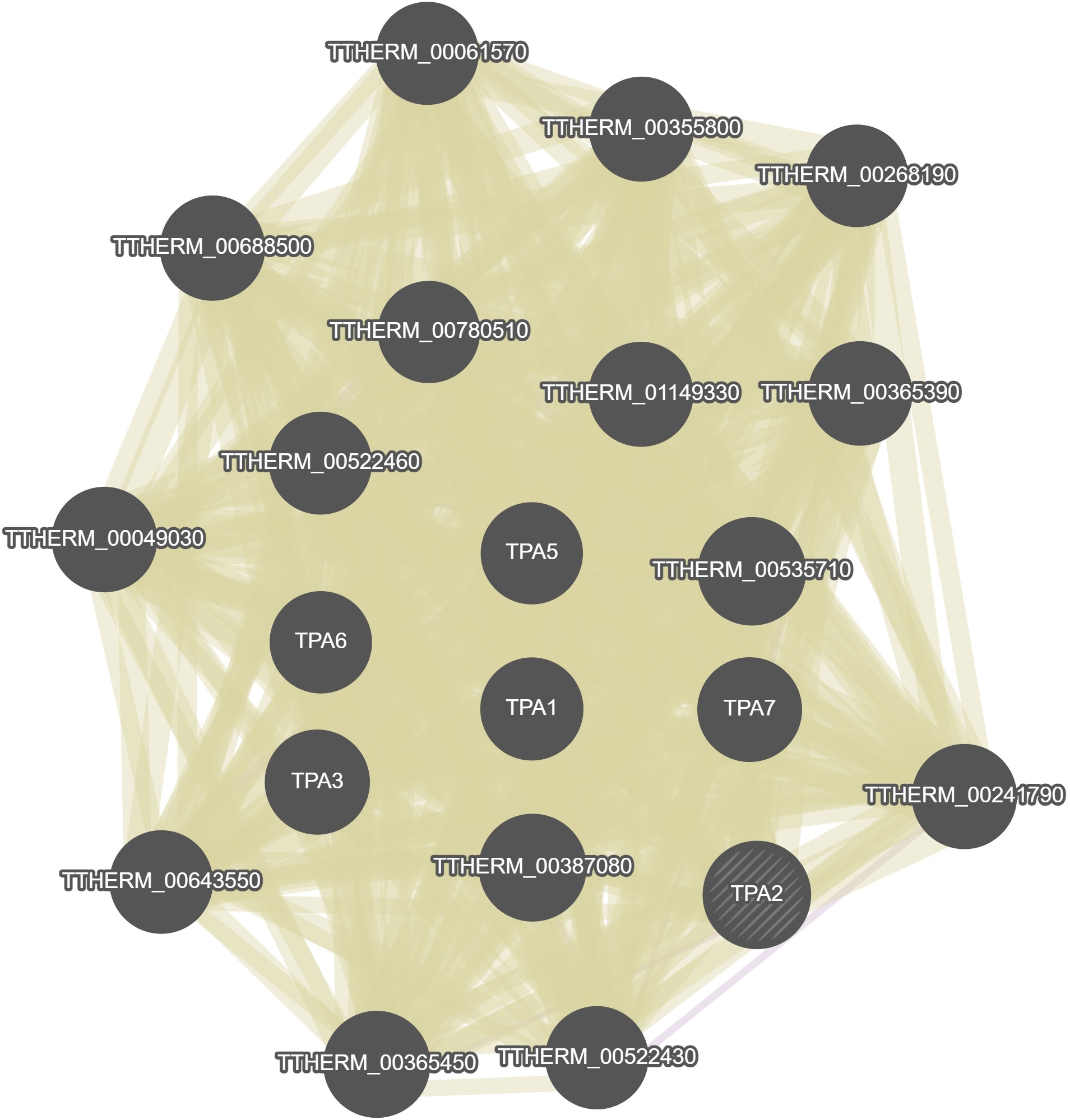
No results were calculated for this gene in GeneMania.
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:11952090: Linder JU, Schultz JE (2002) Guanylyl cyclases in unicellular organisms. Molecular and cellular biochemistry 230(1-2):149-58
- Ref:10428960: Linder JU, Engel P, Reimer A, Krüger T, Plattner H, Schultz A, Schultz JE (1999) Guanylyl cyclases with the topology of mammalian adenylyl cyclases and an N-terminal P-type ATPase-like domain in Paramecium, Tetrahymena and Plasmodium. The EMBO journal 18(15):4222-32
- Ref:9405804: Wang S, Gao D, Penny J, Krishna S, Takeyasu K (1997) P-type ATPases in Tetrahymena. Annals of the New York Academy of Sciences 834( ):158-60
- Ref:9124316: Wang S, Takeyasu K (1997) Primary structure and evolution of the ATP-binding domains of the P-type ATPases in Tetrahymena thermophila. The American journal of physiology 272(2 Pt 1):C715-28
 Sequences
Sequences
>TTHERM_00700900(coding)
ATGAGTTAAGTTAATGTCAGGTAGTCTGCCTCTCTCGTAAGAAAATTCGAAGAAATGAGG
CAGTCTATCTAAATCAACAACTCTATTCAGCAAATGAGATAGAGTTAATAGTACTAGACT
GGTATTCAAGAAAACTAAGATGGCAACAAAGTAGCCCATAGAAACGCAGACGTCCTCGAA
GAAATAGGTTTGAAGACTGAAGATGTCTTCATCAATAACAATGGAACATAGGTATAATAG
CAACAAGATCACCATGCTAAAGGCAAATAAATTTCTAAAGAAGACAAGTCTAAGAAAGAA
TTAGTCGAAAAAGTTGACCACAAGATTTCTCTTGAGGAACTTAGATAAAAGTATGAAACT
GACTACTAAAAGGGTCTTACCGAAGAATAAGCTGCTCATTTACTCAAAATTCATGGTGAG
AATAAACTTACTGAGAAGGTAAAAACTCCTTTCTGGGTTAAGATTCTTATTGAATTGACT
AATGGTTTCGCGCTCTTACTTTGGATCAGTGCTGGTTTATGTTTCCTTGCTTATGGCCTA
AGTCCTGATGATCCCTCTAACATCTATTTGGCTATTGTCATTCTCGTTGTCATCTTCATC
ACTACCGCTATTACTTTCCAACAGAATTCCAAGAGTGAAGCTCTGATGAACTCATTTAAA
AACTTCATTCCTGCAAAATGTATTGTAATCAGAGGAGGATAACCAAAGTCAATTGATGCA
GCTCACTTGGTTGTTGGTGATATAGTATCTATCAGATTGGGAGAGAAGATACCAGCTGAT
ATTCGTATCCTTGAATCAAATGAAATGAAAGTTGATAACTCACCTCTCACTGGTGAGTGC
GAACCTCTTTTGCGTACTGTTGAGTGCTCTCATCCCGAAAGTTATCTAGAAACCTCAAAT
ATTGCTTTCTTTGGTACTTTATGCAAAGAAGGTAATGGCAAAGGTATTGTCATTTGCACC
GGTGATAGAACTACTCTTGGATAAATCGCTGATTTGTCATCAGGTGAGAAAAAGGTTAAG
ACTCCTCTTCGTTAAGAACTTGACCGTTTTGTCATTTTAATTACTATCATCGCTATTTTC
CTTGGTGTTCTTTTCTTCTTGTTAGCCTACTTCTACATGAAATACGATTACATGGTTTGC
ATCGTTTTTGGTATTGGTATTCTTGTCGCTAACGTACCCGAAGGTTTATTGGGTTGCATT
ACCATTTCCTTGGCTATTACTGCTAAGAATCTCGCTGTAAAAAATGTTCTTGTTAAGAAT
CTAGAAGCTGTTGAAACCCTTGGTTCTACTTCTTGTATTTGCTCAGACAAAACTGGTACT
CTTACTCAGAATGTTATGTCTGTCAAGAATATGTGGTTTAAGGATAAGATTTACATGTGC
AAGAACAAAGTCCATCTTAAGCAAGGTGAAATTCCTGAGTACGATATCAACGATAACGAT
TTCTAAACCCTTCAAAAGGCTGCTATGTTGAGTAGTGAAGCTCGTTTTGATACCTCCTCA
GTTAAAGACCAATCTAACATCGACTACATAACATGTCCAGTTATGGGAGATGCCACCGAA
ACTGGTATCATTAGATTCTTTTAATACATTGATGATGTCAACAAATTCCGTGAAAGATAT
TAAATTGCTAAAAACCCTGATGGAACATACGGTAAAATGCCTTTTAACTCATAAGTTAAA
TTTGCATTGACCATCATTCAAGAATAGCTTCCTGGTAGCAACTACACTGTTTATATCAAA
GGTGCTCCTGAAAAGATTTGGAGTTACTGCAATTCAGTTATGATTAATGGCTAACCAAGT
CAAATTGATTAAACATGGTAGAAGAAATTCAAAGCAGTCAATCTCACTTTTGGTAAGGGA
GGTGAAAGAGTTCTTGGATTTGCTAAGCTCCATTTACCTGCTGAAGATTTCCCTGAAGGC
TTTATTTTCAATGTTTCTTCACTTTAAAAATTCCCCTTCAAATTGGCTAATTTCTAGTTC
TGCGGTCTTATTTCATTGATGGATCCTCCTAAAACTAGAGTTCCCTATGCAATTTTGGAA
TGCAGATCTGCTGGTGTCAAAGTTATCATGGTTACAGGCGATCAACCTCCTACTGCTGCT
GCCATCGCCAAAGAAGTCAACATTATTCCTAAGGAAGTTATTACTAATGAAGATATCTTG
GAACAAAATCCCAGTAAAACTTGGTGGGAAGCAAGTGAAGAGTGCGAGGCTATCATTGTC
CATGGTGATAGAATCGTAGAATCGTTTGAGAAATCATTATCTGAGTAAAAATAAGAAAAC
TTCTACTTGCGTTAATGGGTAAAAAAACAATATTGCGTATTTGCAAGAACAACACCTGCT
CAAAAATTACAAATTGTAGATGCTTGCCAAATGGAAGGATTTATTGTCGCTGCAACAGGT
GATGGTGTCAACGACTCTCCTGCTATCAAGAAAGCTGATATTGGTATTTCAATGAACTTG
TCAGGATCTGATGTTACTAAAGATGCTGCTGATATGGTTTTAATTGATGATGATTTTGCT
TCTATTGTTTTGGGTGTTGAAGAAGGTAGAAAAATCTTCGATAACTTAAAGAAGACTGTT
GTCTATCTTCTTACTTCTAATATGACTGAAGTCGTTCCCTTCTTGGCTTTCATTATTTTG
GAATTACCTTTACCTCTCTCTAGTATTTACATGTTAGTTATTTGTGTCGGAACTGATGTT
TGGCCTGCCATTTCTTTAGCATATGAAGAAGCTGAACTTGATGTTATGACTAGAAGACCT
CGTCTAAAATCTGAACATCTTGTTTCTAATAAATTGATTACAATCGCTTACCTTCAAACT
GGTTAAATTGCAAGTGCAGCAGGATTCCTAGGTTACTATGTTGCATTTAACTATTTCGGT
TTCCCAGTCCTTTCATTATTTGGTATGGCTTCTGGATCAGGTTACAAACCACCCAAAAAT
GATTTCAACGAAAGCTACATCAATCCTATCACAAATTAGATAGACAAGAATTACAATACA
GAAATATATAAGATAACAGGAACTACTCGTTGTGACTAATTAAATAAAGATTAACAAGAT
AAATTAGACAAATTAAAATACACTGTAGACTGGTTTACTGTTACTGAAGGTAACTACGAT
TTAAGAAAGACATTTGTAAAATGTGATACAGATTCAGGTATTTTTGTTCCCATTTACAAA
TGGAGTGACTGCGATATTACCTTGAGTAAAAATAAATCACCTATCACTGACTAAACTGCC
TGTTATTCAACTGATGCTTAAAGATATGCATAAAGTGTCTACTTCATCAGTGTTGTCTTA
CTACAATGGTCAAATATCTTTGCTTGCAAGAGTAGATCAACAAGTTACTCAACCACAGCA
TTTAATTCTATTATGATTCATGGTGTCATCTTCGAAACTTGTTTAACCGTTTTCCTTTAA
TACGTCCCAGGAGTATAAGAAGTCTTCGGTGGTAGACCTCTATTCTTCTGGCTCTGGACA
CCAGCCTTAATTTTCAGTATAACTTTACTTGTTTATGATGAATTAAGAAAATTCCTTTGT
AGACATGTTAAGTGGTATTACAAATACTGTTACTGGTGA
>TTHERM_00700900(gene)
TAAATTTAGATAAAAAATATTATAAATTAATAAATACAGCTAAGTTAAATCAATCTTATT
AGCTACTATTTAAGTATTTATAAATGAGTTAAGTTAATGTCAGGTAGTCTGCCTCTCTCG
TAAGAAAATTCGAAGAAATGAGGCAGTCTATCTAAATCAACAACTCTATTCAGCAAATGA
GATAGAGTTAATAGTACTAGACTGGTATTCAAGAAAACTAAGATGGCAACAAAGTAGCCC
ATAGAAACGCAGACGTCCTCGAAGAAATAGGTTTGAAGACTGAAGATGTCTTCATCAATA
ACAATGGAACATAGGTATAATAGCAACAAGATCACCATGCTAAAGGCAAATAAATTTCTA
AAGAAGACAAGTCTAAGAAAGAATTAGTCGAAAAAGTTGACCACAAGATTTCTCTTGAGG
AACTTAGATAAAAGTATGAAACTGACTACTAAAAGGGTCTTACCGAAGAATAAGCTGCTC
ATTTACTCAAAATTCATGGTGAGAATAAACTTACTGAGAAGGTAAAAACTCCTTTCTGGG
TTAAGATTCTTATTGAATTGACTAATGGTTTCGCGCTCTTACTTTGGATCAGTGCTGGTT
TATGTTTCCTTGCTTATGGCCTAAGTCCTGATGATCCCTCTAACATCTATTTGGCTATTG
TCATTCTCGTTGTCATCTTCATCACTACCGCTATTACTTTCCAACAGAATTCCAAGAGTG
AAGCTCTGATGAACTCATTTAAAAACTTCATTCCTGCAAAATGTATTGTAATCAGAGGAG
GATAACCAAAGTCAATTGATGCAGCTCACTTGGTTGTTGGTGATATAGTATCTATCAGAT
TGGGAGAGAAGATACCAGCTGATATTCGTATCCTTGAATCAAATGAAATGAAAGTTGATA
ACTCACCTCTCACTGGTGAGTGCGAACCTCTTTTGCGTACTGTTGAGTGCTCTCATCCCG
AAAGTTATCTAGAAACCTCAAATATTGCTTTCTTTGGTACTTTATGCAAAGAAGGTAATG
GCAAAGGTATTGTCATTTGCACCGGTGATAGAACTACTCTTGGATAAATCGCTGATTTGT
CATCAGGTGAGAAAAAGGTTAAGACTCCTCTTCGTTAAGAACTTGACCGTTTTGTCATTT
TAATTACTATCATCGCTATTTTCCTTGGTGTTCTTTTCTTCTTGTTAGCCTACTTCTACA
TGAAATACGATTACATGGTTTGCATCGTTTTTGGTATTGGTATTCTTGTCGCTAACGTAC
CCGAAGGTTTATTGGGTTGCATTACCATTTCCTTGGCTATTACTGCTAAGAATCTCGCTG
TAAAAAATGTTCTTGTTAAGAATCTAGAAGCTGTTGAAACCCTTGGTTCTACTTCTTGTA
TTTGCTCAGACAAAACTGGTACTCTTACTCAGAATGTTATGTCTGTCAAGAATATGTGGT
TTAAGGATAAGATTTACATGTGCAAGAACAAAGTCCATCTTAAGCAAGGTGAAATTCCTG
AGTACGATATCAACGATAACGATTTCTAAACCCTTCAAAAGGCTGCTATGTTGAGTAGTG
AAGCTCGTTTTGATACCTCCTCAGTTAAAGACCAATCTAACATCGACTACATAACATGTC
CAGTTATGGGAGATGCCACCGAAACTGGTATCATTAGATTCTTTTAATACATTGATGATG
TCAACAAATTCCGTGAAAGATATTAAATTGCTAAAAACCCTGATGGAACATACGGTAAAA
TGCCTTTTAACTCATAAGTTAAATTTGCATTGACCATCATTCAAGAATAGCTTCCTGGTA
GCAACTACACTGTTTATATCAAAGGTGCTCCTGAAAAGATTTGGAGTTACTGCAATTCAG
TTATGATTAATGGCTAACCAAGTCAAATTGATTAAACATGGTAGAAGAAATTCAAAGCAG
TCAATCTCACTTTTGGTAAGGGAGGTGAAAGAGTTCTTGGATTTGCTAAGCTCCATTTAC
CTGCTGAAGATTTCCCTGAAGGCTTTATTTTCAATGTTTCTTCACTTTAAAAATTCCCCT
TCAAATTGGCTAATTTCTAGTTCTGCGGTCTTATTTCATTGATGGATCCTCCTAAAACTA
GAGTTCCCTATGCAATTTTGGAATGCAGATCTGCTGGTGTCAAAGTTATCATGGTTACAG
GCGATCAACCTCCTACTGCTGCTGCCATCGCCAAAGAAGTCAACATTATTCCTAAGGAAG
TTATTACTAATGAAGATATCTTGGAACAAAATCCCAGTAAAACTTGGTGGGAAGCAAGTG
AAGAGTGCGAGGCTATCATTGTCCATGGTGATAGAATCGTAGAATCGTTTGAGAAATCAT
TATCTGAGTAAAAATAAGAAAACTTCTACTTGCGTTAATGGGTAAAAAAACAATATTGCG
TATTTGCAAGAACAACACCTGCTCAAAAATTACAAATTGTAGATGCTTGCCAAATGGAAG
GATTTATTGTCGCTGCAACAGGTGATGGTGTCAACGACTCTCCTGCTATCAAGAAAGCTG
ATATTGGTATTTCAATGAACTTGTCAGGATCTGATGTTACTAAAGATGCTGCTGATATGG
TTTTAATTGATGATGATTTTGCTTCTATTGTTTTGGGTGTTGAAGAAGGTAGAAAAATCT
TCGATAACTTAAAGAAGACTGTTGTCTATCTTCTTACTTCTAATATGACTGAAGTCGTTC
CCTTCTTGGCTTTCATTATTTTGGAATTACCTTTACCTCTCTCTAGTATTTACATGTTAG
TTATTTGTGTCGGAACTGATGTTTGGCCTGCCATTTCTTTAGCATATGAAGAAGCTGAAC
TTGATGTTATGACTAGAAGACCTCGTCTAAAATCTGAACATCTTGTTTCTAATAAATTGA
TTACAATCGCTTACCTTCAAACTGGTTAAATTGCAAGTGCAGCAGGATTCCTAGGTTACT
ATGTTGCATTTAACTATTTCGGTTTCCCAGTCCTTTCATTATTTGGTATGGCTTCTGGAT
CAGGTTACAAACCACCCAAAAATGATTTCAACGAAAGCTACATCAATCCTATCACAAATT
AGATAGACAAGAATTACAATACAGAAATATATAAGATAACAGGAACTACTCGTTGTGACT
AATTAAATAAAGATTAACAAGATAAATTAGACAAATTAAAATACACTGTAGACTGGTTTA
CTGTTACTGAAGGTAACTACGATTTAAGAAAGACATTTGTAAAATGTGATACAGATTCAG
GTATTTTTGTTCCCATTTACAAATGGAGTGACTGCGATATTACCTTGAGTAAAAATAAAT
CACCTATCACTGACTAAACTGCCTGTTATTCAACTGATGCTTAAAGATATGCATAAAGTG
TCTACTTCATCAGTGTTGTCTTACTACAATGGTCAAATATCTTTGCTTGCAAGAGTAGAT
CAACAAGTTACTCAACCACAGCATTTAATTCTATTATGATTCATGGTGTCATCTTCGAAA
CTTGTTTAACCGTTTTCCTTTAATACGTCCCAGGAGTATAAGAAGTCTTCGGTGGTAGAC
CTCTATTCTTCTGGCTCTGGACACCAGCCTTAATTTTCAGTATAACTTTACTTGTTTATG
ATGAATTAAGAAAATTCCTTTGTAGACATGTTAAGTGGTATTACAAATACTGTTACTGGT
GAAAAAAATATAATTCACTAAGATATAAAGAAAGAAAGTCTATTAATTCTTAAATTAATC
ATTTAATATTACTATAAAATAATAAATAAATATATATATCTATCTCCATTTTTGTTTAAT
AATTTAAATACATAAATAAATTAATAAATGTTACTTCGATATATTTTGTATTTTCTTAAC
ATTATCCTTGATTGCTTAACTTAAAATTCATAATTAAAATACGAGATTGCATTTTTTAAG
TAATATTTGCTTATCTTTTACAATATT
>TTHERM_00700900(protein)
MSQVNVRQSASLVRKFEEMRQSIQINNSIQQMRQSQQYQTGIQENQDGNKVAHRNADVLE
EIGLKTEDVFINNNGTQVQQQQDHHAKGKQISKEDKSKKELVEKVDHKISLEELRQKYET
DYQKGLTEEQAAHLLKIHGENKLTEKVKTPFWVKILIELTNGFALLLWISAGLCFLAYGL
SPDDPSNIYLAIVILVVIFITTAITFQQNSKSEALMNSFKNFIPAKCIVIRGGQPKSIDA
AHLVVGDIVSIRLGEKIPADIRILESNEMKVDNSPLTGECEPLLRTVECSHPESYLETSN
IAFFGTLCKEGNGKGIVICTGDRTTLGQIADLSSGEKKVKTPLRQELDRFVILITIIAIF
LGVLFFLLAYFYMKYDYMVCIVFGIGILVANVPEGLLGCITISLAITAKNLAVKNVLVKN
LEAVETLGSTSCICSDKTGTLTQNVMSVKNMWFKDKIYMCKNKVHLKQGEIPEYDINDND
FQTLQKAAMLSSEARFDTSSVKDQSNIDYITCPVMGDATETGIIRFFQYIDDVNKFRERY
QIAKNPDGTYGKMPFNSQVKFALTIIQEQLPGSNYTVYIKGAPEKIWSYCNSVMINGQPS
QIDQTWQKKFKAVNLTFGKGGERVLGFAKLHLPAEDFPEGFIFNVSSLQKFPFKLANFQF
CGLISLMDPPKTRVPYAILECRSAGVKVIMVTGDQPPTAAAIAKEVNIIPKEVITNEDIL
EQNPSKTWWEASEECEAIIVHGDRIVESFEKSLSEQKQENFYLRQWVKKQYCVFARTTPA
QKLQIVDACQMEGFIVAATGDGVNDSPAIKKADIGISMNLSGSDVTKDAADMVLIDDDFA
SIVLGVEEGRKIFDNLKKTVVYLLTSNMTEVVPFLAFIILELPLPLSSIYMLVICVGTDV
WPAISLAYEEAELDVMTRRPRLKSEHLVSNKLITIAYLQTGQIASAAGFLGYYVAFNYFG
FPVLSLFGMASGSGYKPPKNDFNESYINPITNQIDKNYNTEIYKITGTTRCDQLNKDQQD
KLDKLKYTVDWFTVTEGNYDLRKTFVKCDTDSGIFVPIYKWSDCDITLSKNKSPITDQTA
CYSTDAQRYAQSVYFISVVLLQWSNIFACKSRSTSYSTTAFNSIMIHGVIFETCLTVFLQ
YVPGVQEVFGGRPLFFWLWTPALIFSITLLVYDELRKFLCRHVKWYYKYCYW
 Identifiers and Description
Identifiers and Description
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center  Homologs
Homologs
 General Information
General Information
 Associated Literature
Associated Literature
 Sequences
Sequences



