Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00723170Standard Name
RAB36 (RAB GTPase 36)Aliases
PreTt18077 | 116.m00102 | 3671.m00026Description
RAB36 small guanosine triphosphatase family Ras family protein; Novel Rab family protein; small monomeric GTPase that regulates membrane fusion and fission events; Small GTPaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- catalase complex (IEA) | GO:0062151
- lysosomal HOPS complex (IEA) | GO:1902501
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- sigma factor antagonist complex (IEA) | GO:1903865
Molecular Function
- 1-deoxy-D-xylulose-5-phosphate synthase activity (IEA) | GO:0008661
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- amorpha-4,11-diene 12-monooxygenase activity (IEA) | GO:0062150
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- insulin-like growth factor II binding (IEA) | GO:0031995
- isochorismate synthase activity (IEA) | GO:0008909
- kanamycin kinase activity (IEA) | GO:0008910
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- microtubule severing ATPase activity (IEA) | GO:0008568
- mRNA (cytidine-5-)-methyltransferase activity (IEA) | GO:0062152
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete gamma-aminovinylacetate deaminase activity (IEA) | GO:0034886
- obsolete glucan endo-1,4-beta-glucanase activity, C-3 substituted reducing group (IEA) | GO:0052862
- obsolete isonicotinic acid hydrazide hydrolase activity (IEA) | GO:0034876
- obsolete myosin ATPase activity (IEA) | GO:0008570
- obsolete peroxisome-assembly ATPase activity (IEA) | GO:0008573
- obsolete serpin (IEA) | GO:0004868
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylinositol-3-phosphate binding (IEA) | GO:0032266
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sulfoquinovose isomerase activity (IEA) | GO:0061593
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- type 7 serotonin receptor binding (IEA) | GO:0031833
- zinc efflux active transmembrane transporter activity (IEA) | GO:0015341
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- allantoate transport (IEA) | GO:0015719
- cellular response to organic cyclic compound (IEA) | GO:0071407
- cellular response to vanadate(3-) (IEA) | GO:1902439
- DNA replication preinitiation complex assembly (IEA) | GO:0071163
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- homogentisate metabolic process (IEA) | GO:1901999
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-19 production (IEA) | GO:0032622
- interleukin-2 production (IEA) | GO:0032623
- interleukin-28A secretion (IEA) | GO:0072628
- lipid homeostasis (IEA) | GO:0055088
- modulation by organism of innate immune response in other organism involved in symbiotic interaction (IEA) | GO:0052306
- mucosal tolerance induction (IEA) | GO:0002427
- negative regulation of DNA amplification (IEA) | GO:1904524
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete induction by organism of defense-related symbiont reactive oxygen species production (IEA) | GO:0052401
- obsolete peptidyl-lysine deglutarylation (IEA) | GO:0061699
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- phosphorelay signal transduction system (IEA) | GO:0000160
- plant-type cell wall modification (IEA) | GO:0009827
- polyamine acetylation (IEA) | GO:0032917
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antifungal innate immune response (IEA) | GO:1905036
- positive regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002600
- positive regulation of deacetylase activity (IEA) | GO:0090045
- positive regulation of I-kappaB phosphorylation (IEA) | GO:1903721
- positive regulation of protein autoubiquitination (IEA) | GO:1902499
- positive regulation of turning behavior involved in mating (IEA) | GO:0061095
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to chloroplast (IEA) | GO:0072598
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002598
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of hyphopodium formation (IEA) | GO:0075188
- regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process (IEA) | GO:1902962
- regulation of mRNA 3'-UTR binding (IEA) | GO:1903837
- regulation of protein localization to cell cortex (IEA) | GO:1904776
- regulation of stress response to copper ion (IEA) | GO:1903853
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- response to curcumin (IEA) | GO:1904643
- response to ethylene (IEA) | GO:0009723
- response to osmotic stress (IEA) | GO:0006970
- response to oxidative stress (IEA) | GO:0006979
- striated muscle contraction (IEA) | GO:0006941
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- thymidine catabolic process (IEA) | GO:0006214
 Domains
Domains
- ( PF00071 ) Ras family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
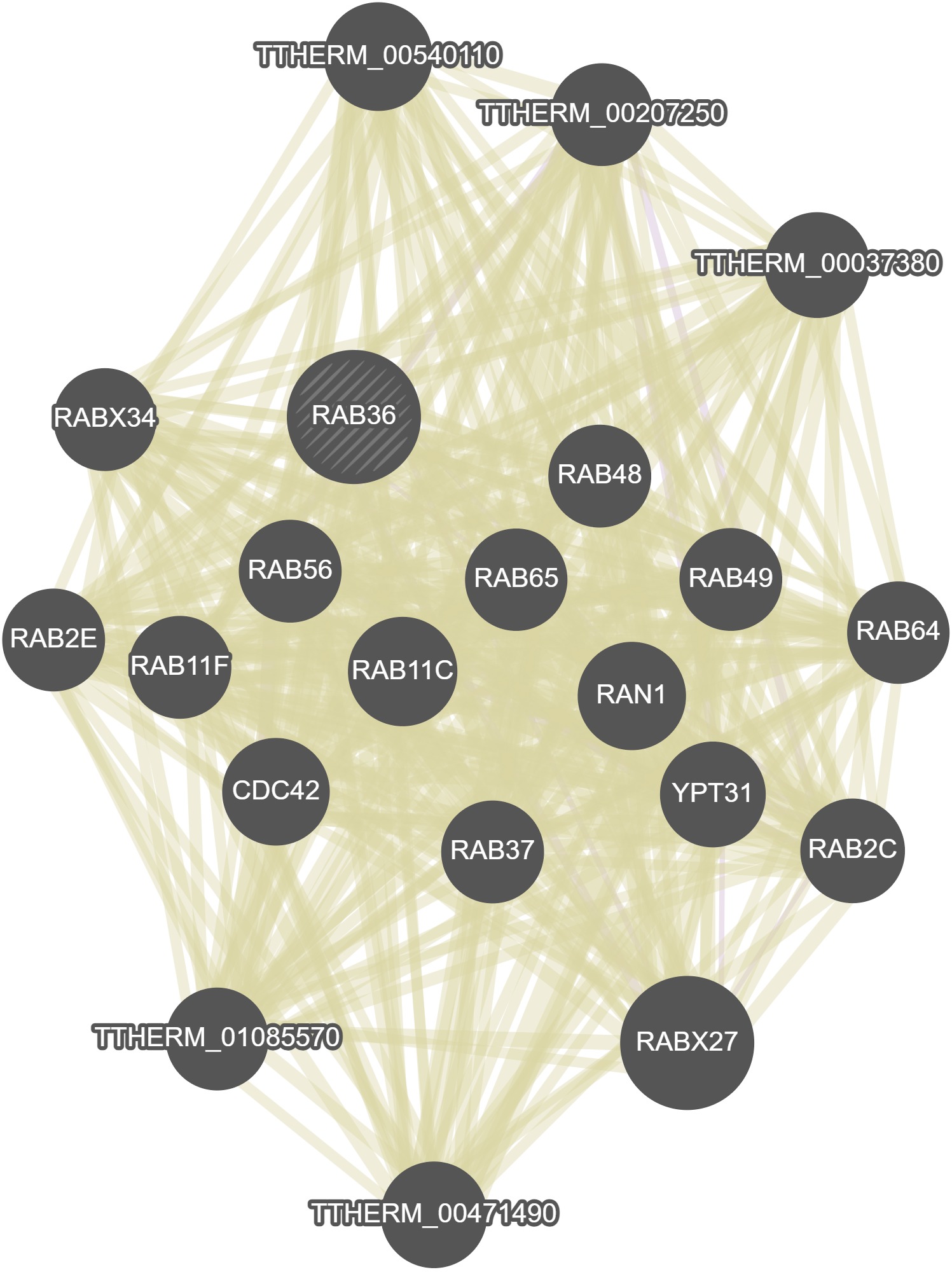
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_00723170(coding)
ATGGCAGCATTATAAAAAAAGCAATCCATAAACTGTAAAGTGTTAGTTTTAGGTAAATCA
GGAGTTGGAAAGAGTTATATTCTTCTTTAATATACAGCTCCTCCTCAAGAAATAGTCTAG
ATGCCAAAATTAACAATTGGTGTAGATTATAAATATGGGACTGAAAAGATTGAAGATACA
GAAGTTAAAATGTCAATATGGGATACTGCTGGCCAAGAGAAATATCGCCCTATTATTTAG
ACTTATTTTAAGAATTCAAAGGCAGCAATTCTTGTGTTTGATTTAACAGATAAGTAATCC
TTGTAAGATGTGAGATACTGGCTAAGAGAAGTAAAATTGTAATGTGGTAAGGATGTAGCA
AAAATTTTAGTAGGAAATAAATGTGATTTGCCTCACGAATAGCCTGATTAAGCTTTGATA
TAAGAATATTGCGAAGAATTTCAAATGGAATATATAGAATCAAGTGCTTTATAAAAGATT
AATATTTAAGAAATATTTGTAAAGTTAGCTAAAAAGATATATTAATAGCACAAAGCAAGT
ATTCCATAAAATCAATTAAAATAAAATAGTTTACCAGCAAATGAAGCTTAAAATTAAAAT
TCTACATCATCAAATAAATAAATAAGTTTAAAAAATTCTAAACCAAAATAGAAAAAAGAA
AATGATGGGTGTTGCTGA>TTHERM_00723170(gene)
AATTAAATAAAAAAATCTGATTAAACTCACAACTTAGCAGCTTTGATAGAAACTAATTTT
TAGAAATGGCAGCATTATAAAAAAAGCAATCCATAAACTGTAAAGTGTTAGTTTTAGGTA
AATCAGGAGTTGGAAAGAGTTATATTCTTCTTTAATATACAGCTCCTCCTCAAGAAATAG
TCTAGATGCCAAAATTAACAATTGGTGTAGATTATAAATATGGGACTGAAAAGATTGAAG
ATACAGAAGTTAAAATGTCAATATGGGATACTGCTGGCCAAGAGAAATATCGCCCTATTA
TTTAGACTTATTTTAAGAATTCAAAGGCAGCAATTCTTGTGTTTGATTTAACAGATAAGT
AATCCTTGTAAGATGTGAGATACTGGCTAAGAGAAGTAAAATTGTAATGTGGTAAGGATG
TAGCAAAAATTTTAGTAGGAAATAAATGTGATTTGCCTCACGAATAGCCTGATTAAGCTT
TGATATAAGAATATTGCGAAGAATTTCAAATGGAATATATAGAATCAAGTGCTTTATAAA
AGATTAATATTTAAGAAATATTTGTAAAGTTAGCTAAAAAGATATATTAATAGCACAAAG
CAAGTATTCCATAAAATCAATTAAAATAAAATAGTTTACCAGCAAATGAAGCTTAAAATT
AAAATTCTACATCATCAAATAAATAAATAAGTTTAAAAAATTCTAAACCAAAATAGAAAA
AAGAAAATGATGGGTGTTGCTGATGAAATATGTTCATTTATTCAACCATATTTATTCAGA
GCAAATCTAAGCAAATATTTCTGTGAGATAAAAAAATTAATAAAAGCAAATTCTTTAATT
TGTAAAAATATTTCTAAATTATATTCTAAAATAATATTTTACTAACAAAAATAATTTTTA
ATAAACTAAATTTAACTATCACGAAAGTGTGTAAATAATTTATATTAATTTCAGAATGTT
TAATATTTACATAGAATAATTGTCTATTAAAAATGCATTTTTATCAAATAAATAATTTAA
ATTAAA>TTHERM_00723170(protein)
MAALQKKQSINCKVLVLGKSGVGKSYILLQYTAPPQEIVQMPKLTIGVDYKYGTEKIEDT
EVKMSIWDTAGQEKYRPIIQTYFKNSKAAILVFDLTDKQSLQDVRYWLREVKLQCGKDVA
KILVGNKCDLPHEQPDQALIQEYCEEFQMEYIESSALQKINIQEIFVKLAKKIYQQHKAS
IPQNQLKQNSLPANEAQNQNSTSSNKQISLKNSKPKQKKENDGCC