Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00732590Standard Name
EIF6 (Eukaryotic translation Initiation Factor 6)Aliases
PreTt02780 | 119.m00105Description
EIF6 eukaryotic translation initiation factor 6; Translation initiation factor eIF-6, putative family protein; Translation initiation factor IF6Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- ionotropic glutamate receptor complex (IEA) | GO:0008328
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- CCR7 chemokine receptor binding (IEA) | GO:0031732
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- fatty-acyl-CoA binding (IEA) | GO:0000062
- ferric-chelate reductase (NADPH) activity (IEA) | GO:0052851
- G protein-coupled peptide receptor activity (IEA) | GO:0008528
- medium-chain-(S)-2-hydroxy-acid oxidase activity (IEA) | GO:0052854
- obsolete phospholipid scrambling (IEA) | GO:0008498
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete sulfur-containing amino acid secondary active transmembrane transporter activity (IEA) | GO:1901680
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- proteinase activated receptor binding (IEA) | GO:0031871
- replication fork barrier binding (IEA) | GO:0031634
- type 2 proteinase activated receptor binding (IEA) | GO:0031873
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- bile acid metabolic process (IEA) | GO:0008206
- box H/ACA sno(s)RNA 3'-end processing (IEA) | GO:0000495
- calcium ion export (IEA) | GO:1901660
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cell-cell adhesion involved in sealing an epithelial fold (IEA) | GO:0060607
- cellular response to diacyl bacterial lipopeptide (IEA) | GO:0071726
- cellular response to dopamine (IEA) | GO:1903351
- cleavage between SSU-rRNA and 5.8S rRNA of tetracistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, 2S rRNA, LSU-rRNA) (IEA) | GO:0000484
- coenzyme A metabolic process (IEA) | GO:0015936
- commissural neuron axon guidance (IEA) | GO:0071679
- copulation (IEA) | GO:0007620
- embryonic brain development (IEA) | GO:1990403
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- learning or memory (IEA) | GO:0007611
- mating (IEA) | GO:0007618
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of glycine secretion, neurotransmission (IEA) | GO:1904625
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete autophagic death (sensu Saccharomyces) (IEA) | GO:0007577
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of cGMP catabolic process (IEA) | GO:0030830
- obsolete phosphorylation of RNA polymerase II C-terminal domain (IEA) | GO:0070816
- obsolete regulation of cellular carbohydrate catabolic process (IEA) | GO:0043471
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- obsolete RING-like zinc finger domain-mediated complex assembly (IEA) | GO:0071536
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of male mating behavior (IEA) | GO:1902437
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- pre-replicative complex assembly involved in cell cycle DNA replication (IEA) | GO:1902299
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine nucleobase fermentation (IEA) | GO:0043466
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of male mating behavior (IEA) | GO:1902435
- regulation of phosphorus utilization (IEA) | GO:0006795
- response to methamphetamine hydrochloride (IEA) | GO:1904313
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- vesicle coating (IEA) | GO:0006901
 Domains
Domains
- ( PF01912 ) eIF-6 family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
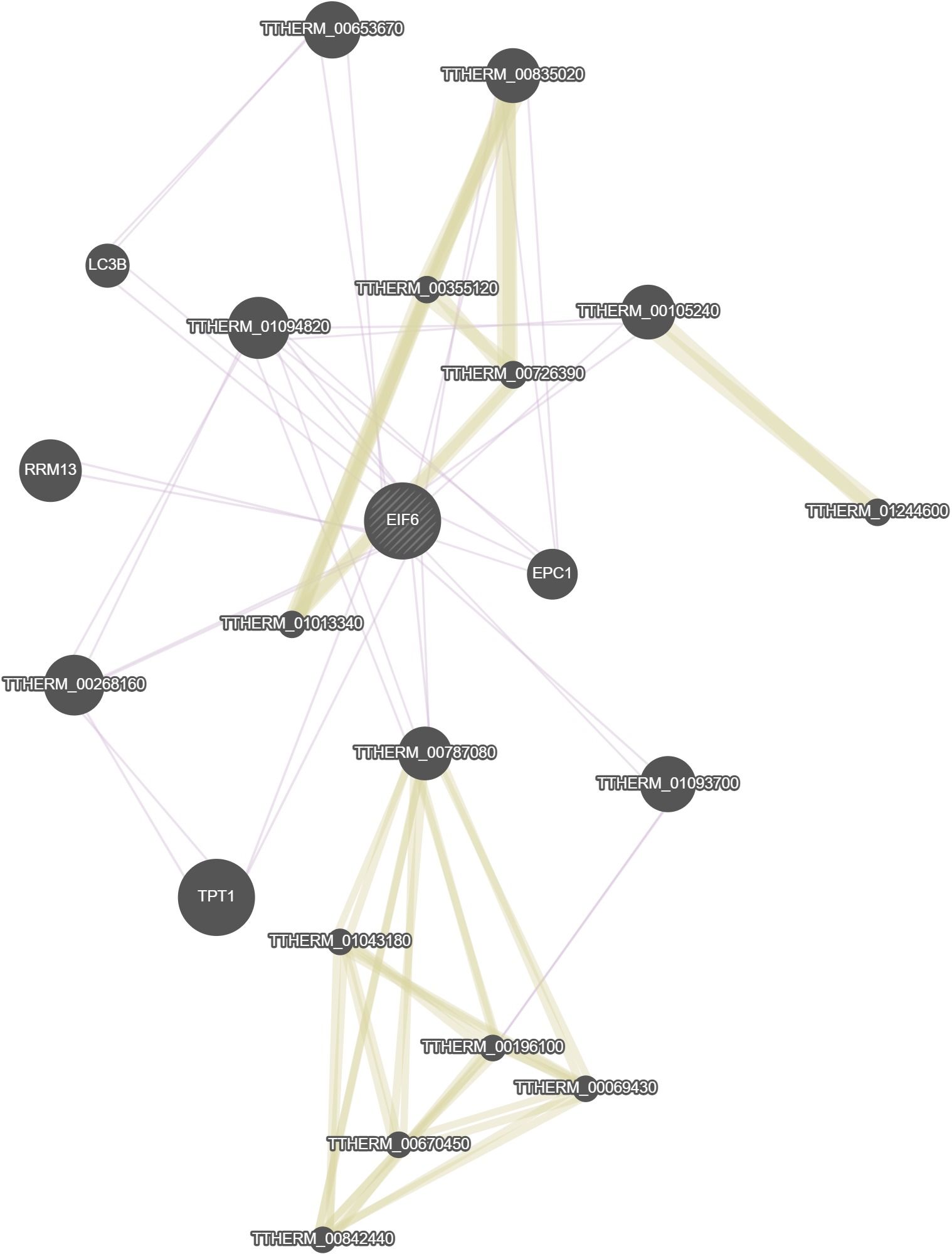
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00732590(coding)
ATGGCAAGAAGATGTTAATTTGAAAACTCCAACGACATCGGAGTCTTCTGCAAACTCACA
TCAGCATACTGTTTAGTTTCCGTTGGTGCATCTGAAAACTTCTATTCTGTTTTTGAATCT
GAATTGGTCCCCCATATTCCAGTTATTCATACATCAATAGGCGGTACTCGTATCGTAGGC
AGAGTAACTTGTGGTAACAAGAACGGTTTGTTGGTCCCTAACACTTGTAATGACAATGAA
TTGAGAAATATTAGAAATTCCCTTCCTGACAACGTCAGAGTAAGAAGAATCGAAGAAAAG
CTTTCAGCTTTAGGTAATTGTGTTGTTGCTAACGATTATGTTGCCTTGATCCACCCTGAT
TTGGACAGAGAAAGTGAAGAAATTATTGCTGACACACTCGGAGTTGAAGTCTTTAGAACT
ACTATTGCTAATAACGTTCTCGTTGGTACTTACTGTGTTATCAATAATAGAGGTGGTCTT
GTTCATCCTTTAGCTTCAGTTGAAGAGTTAGACGAACTTGCTAACCTCCTTTAAATCCCC
CTTTGTGCTGGTACAATTAACAGAGGTTCTGATGTTATTGGTGCTGGCCTCGTTGTAAAT
GATTGGGCTGCTTTTTGTGGTTTAGATACTACTTCTACTGAAATCAGTGTTGTTGAAAAT
ATATTTAAATTAAATGAAATGAAAGATGAAAATATGGATAATGAAATGAGAAAAGACTTC
GTTATGAACCTCGAGTGA>TTHERM_00732590(gene)
CTATTAATTTTGAGGAGTTGATAGCAATCTTGAAGCATATAAAAAAAATTTTAATTTTAT
ATTAAAGAGTCAAAAAAATAAAAAGTAAAGAAAAAAGATAAAAAGAGACAGTCATATAAT
CAAAGTAATTAGAAAGAAATGGCAAGAAGATGTTAATTTGAAAACTCCAACGACATCGGA
GTCTTCTGCAAACTCACATCAGCATACTGTTTAGTTTCCGTTGGTGCATCTGAAAACTTC
TATTCTGTTTTTGAATCTGAATTGGTCCCCCATATTCCAGTTATTCATACATCAATAGGC
GGTACTCGTATCGTAGGCAGAGTAACTTGTGGTAACAAGAACGGTTTGTTGGTCCCTAAC
ACTTGTAATGACAATGAATTGAGAAATATTAGAAATTCCCTTCCTGACAACGTCAGAGTA
AGAAGAATCGAAGAAAAGCTTTCAGCTTTAGGTAATTGTGTTGTTGCTAACGATTATGTT
GCCTTGATCCACCCTGATTTGGACAGAGAAAGTGAAGAAATTATTGCTGACACACTCGGA
GTTGAAGTCTTTAGAACTACTATTGCTAATAACGTTCTCGTTGGTACTTACTGTGTTATC
AATAATAGAGGTGGTCTTGTTCATCCTTTAGCTTCAGTTGAAGAGTTAGACGAACTTGCT
AACCTCCTTTAAATCCCCCTTTGTGCTGGTACAATTAACAGAGGTTCTGATGTTATTGGT
GCTGGCCTCGTTGTAAATGATTGGGCTGCTTTTTGTGGTTTAGATACTACTTCTACTGAA
ATCAGTGTTGTTGAAAATATATTTAAATTAAATGAAATGAAAGATGAAAATATGGATAAT
GAAATGAGAAAAGACTTCGTTATGAACCTCGAGTGAAAGAGTTAGTAATAAGGAGTCAAA
TTGTATTGAACATCTATCTATCTATTAGTTTATTTTCAAAGCAAACTGACTTAATTTAAA
AATGAATGAAGTGTATTATATTTTGGGCCCTTTATATTTCTAAACATATAATTTTACTGA
AAAAAATAAATATTTAAAATTTTCTTTAGCAAGTTAATTAATTAATGTCTTCAAACAAAT
AATTCTTTATAGAGTTTTTTGTTGCAATAGGCCAAAAGAAGACAAAAAATAACTAACAAA
CCACACAAATAAATACCCTAAGAAATTAATATTAATTTATACTTTATATTTTTATATATA
TATAAATATTGTACAATAGAATATATTGTTCTTTT>TTHERM_00732590(protein)
MARRCQFENSNDIGVFCKLTSAYCLVSVGASENFYSVFESELVPHIPVIHTSIGGTRIVG
RVTCGNKNGLLVPNTCNDNELRNIRNSLPDNVRVRRIEEKLSALGNCVVANDYVALIHPD
LDRESEEIIADTLGVEVFRTTIANNVLVGTYCVINNRGGLVHPLASVEELDELANLLQIP
LCAGTINRGSDVIGAGLVVNDWAAFCGLDTTSTEISVVENIFKLNEMKDENMDNEMRKDF
VMNLE