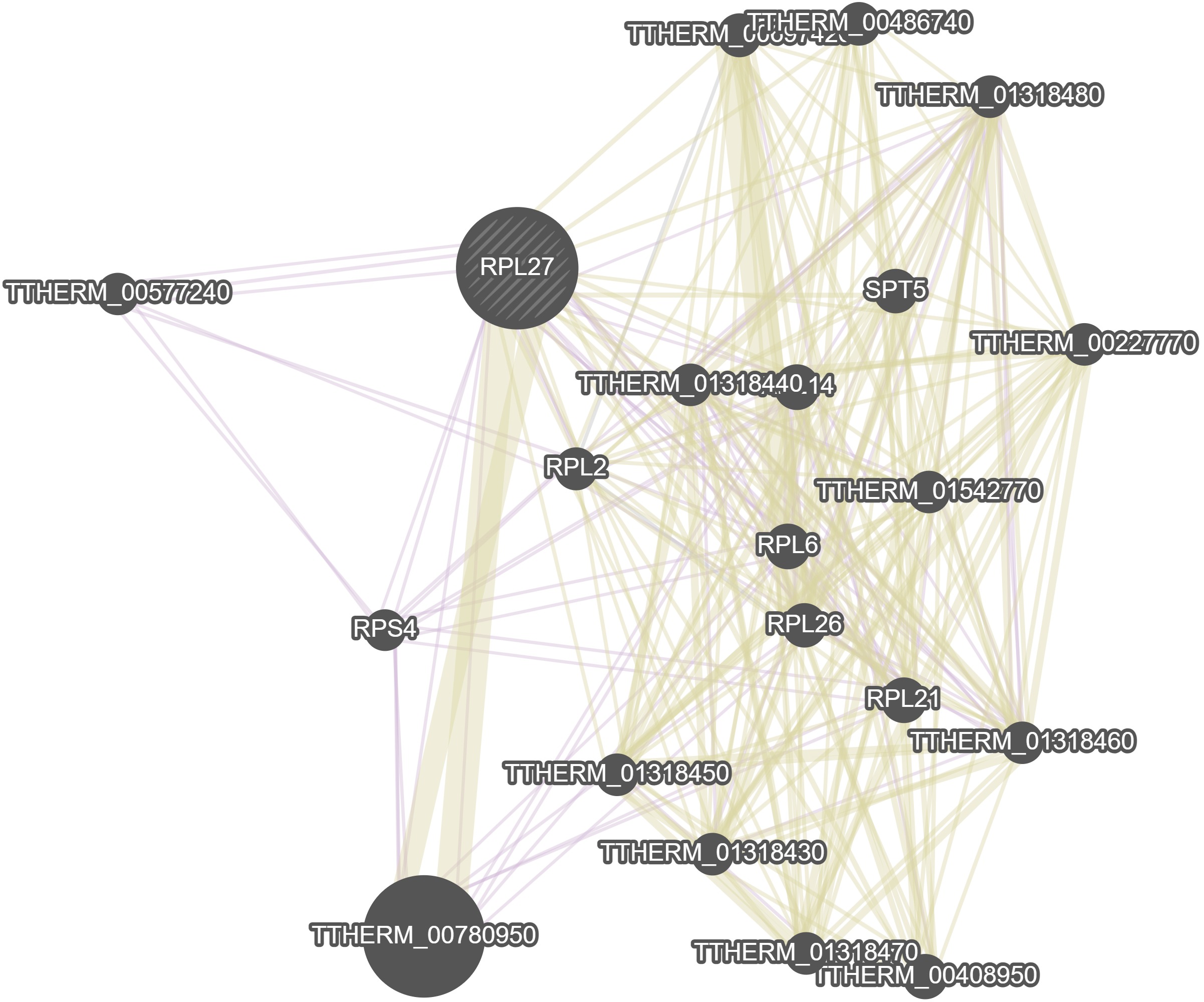
Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00780960Standard Name
RPL27 (Ribosomal Protein of the Large subunit #27)Aliases
PreTt01308 | 130.m00118 | 3738.m00054Description
RPL27 60S ribosomal protein L27; Homolog of Yeast RPL27, Human RPL27; Alias RPL27e; Large ribosomal subunit protein eL27Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha6-beta1 integrin-CD151 complex (IEA) | GO:0071059
- catalase complex (IEA) | GO:0062151
- ionotropic glutamate receptor complex (IEA) | GO:0008328
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 11beta-hydroxyprogesterone binding (IEA) | GO:1903879
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- adiponectin binding (IEA) | GO:0055100
- amino acid transmembrane transporter activity (IEA) | GO:0015171
- CCR10 chemokine receptor binding (IEA) | GO:0031735
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- ferric-chelate reductase (NADPH) activity (IEA) | GO:0052851
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- ion channel inhibitor activity (IEA) | GO:0008200
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- medium-chain-(S)-2-hydroxy-acid oxidase activity (IEA) | GO:0052854
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete phospholipid scrambling (IEA) | GO:0008498
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- peptide-glutamate-alpha-N-acetyltransferase activity (IEA) | GO:1990190
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- protein histidine kinase activity (IEA) | GO:0004673
- proteinase activated receptor binding (IEA) | GO:0031871
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- replication fork barrier binding (IEA) | GO:0031634
- synthetic cannabinoid binding (IEA) | GO:1904483
- type 1 proteinase activated receptor binding (IEA) | GO:0031872
- type 2 proteinase activated receptor binding (IEA) | GO:0031873
Biological Process
- adult walking behavior (IEA) | GO:0007628
- androgen metabolic process (IEA) | GO:0008209
- asymmetric protein localization (IEA) | GO:0008105
- basement membrane organization (IEA) | GO:0071711
- C21-steroid hormone catabolic process (IEA) | GO:0008208
- C21-steroid hormone metabolic process (IEA) | GO:0008207
- calcium ion export (IEA) | GO:1901660
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to diacyl bacterial lipopeptide (IEA) | GO:0071726
- coenzyme A biosynthetic process (IEA) | GO:0015937
- coenzyme A metabolic process (IEA) | GO:0015936
- commissural neuron axon guidance (IEA) | GO:0071679
- D-tagatose 6-phosphate catabolic process (IEA) | GO:2001059
- embryonic brain development (IEA) | GO:1990403
- evasion or tolerance by organism of reactive oxygen species produced by other organism involved in symbiotic interaction (IEA) | GO:0052385
- external genitalia morphogenesis (IEA) | GO:0035261
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- hair cycle process (IEA) | GO:0022405
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- homogentisate metabolic process (IEA) | GO:1901999
- humoral immune response (IEA) | GO:0006959
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- induction by organism of defense-related reactive oxygen species production in other organism involved in symbiotic interaction (IEA) | GO:0052264
- learning or memory (IEA) | GO:0007611
- long-term memory (IEA) | GO:0007616
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- mating behavior (IEA) | GO:0007617
- microtubule depolymerization (IEA) | GO:0007019
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation by organism of jasmonic acid-mediated defense response of other organism involved in symbiotic interaction (IEA) | GO:0052266
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of antimicrobial peptide secretion (IEA) | GO:0002795
- negative regulation of bacterial-type flagellum-dependent cell motility (IEA) | GO:1902201
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of glycine secretion, neurotransmission (IEA) | GO:1904625
- negative regulation of glycoprotein metabolic process (IEA) | GO:1903019
- negative regulation of integrin-mediated signaling pathway (IEA) | GO:2001045
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of response to drug (IEA) | GO:2001024
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of stomatal opening (IEA) | GO:1902457
- negative regulation of telomere single strand break repair (IEA) | GO:1903824
- nucleotide-binding oligomerization domain containing 2 signaling pathway (IEA) | GO:0070431
- obsolete activation of MAPK during sporulation (sensu Saccharomyces) (IEA) | GO:0000205
- obsolete autophagic death (sensu Saccharomyces) (IEA) | GO:0007577
- obsolete bent DNA binding (IEA) | GO:0006326
- obsolete larval behavior (sensu Insecta) (IEA) | GO:0007627
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete negative regulation of cGMP catabolic process (IEA) | GO:0030830
- obsolete nucleolar fragmentation involved in replicative aging (IEA) | GO:0001303
- obsolete positive regulation of diuresis by angiotensin (IEA) | GO:0003102
- obsolete positive regulation of mitotic G1 cell cycle arrest in response to nitrogen starvation (IEA) | GO:1903694
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete regulation of telomeric RNA transcription from RNA pol II promoter (IEA) | GO:1901580
- obsolete ribonucleoprotein complex import into nucleus (IEA) | GO:0071167
- obsolete RING-like zinc finger domain-mediated complex assembly (IEA) | GO:0071536
- ommochrome biosynthetic process (IEA) | GO:0006727
- palisade mesophyll development (IEA) | GO:1903866
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- pigment accumulation in response to UV light (IEA) | GO:0043478
- pigment accumulation in tissues in response to UV light (IEA) | GO:0043479
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antifungal peptide biosynthetic process (IEA) | GO:0006967
- positive regulation of B cell mediated immunity (IEA) | GO:0002714
- positive regulation of biosynthetic process of antibacterial peptides active against Gram-positive bacteria (IEA) | GO:0006965
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of turning behavior involved in mating (IEA) | GO:0061095
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- post-chaperonin tubulin folding pathway (IEA) | GO:0007023
- pre-replicative complex assembly involved in cell cycle DNA replication (IEA) | GO:1902299
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine nucleobase fermentation (IEA) | GO:0043466
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antisense RNA transcription (IEA) | GO:0060194
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of dendrite extension (IEA) | GO:1903859
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of glycoprotein metabolic process (IEA) | GO:1903018
- regulation of macrophage derived foam cell differentiation (IEA) | GO:0010743
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903689
- sesquiterpenoid biosynthetic process (IEA) | GO:0016106
- sodium-independent thromboxane transport (IEA) | GO:0071721
- sperm aster formation (IEA) | GO:0035044
- synaptic vesicle priming (IEA) | GO:0016082
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- urethra epithelium development (IEA) | GO:0061071
 Domains
Domains
- ( PF01777 ) Ribosomal L27e protein family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00780960(coding)
ATGGCCAAATTCCTCAAGTACGGTAGAGTTGTCATCCTTTTACAAGGTAGATTCGCTGGT
AAGAAGGCTGTCATCGTCAAGTCCTCTGAAGATGGTACCAAGGACAGAAAGTTCGGTCAC
GTCTTAGTTGCTGGTGTTGAAAGATCCCCCAAGAAGGTCACCAAGAGAATGGGTTCCAAG
AAGATCTAAAAGAGAACTTCCGTCAAGCCCTTCATTAAATACGTCAACCTTAACCATATC
ATGCCCACCAGATACTCCGTTAAGGAACTCTGTGACTTCAAGGAACTTGTTAAGGAAGAT
AAGATCAAGAACAACGCTAAGTCTGAAGTTAGAGACACCTTAAAGAAGGTCTTCGTCGAA
AAGTACAGAACCATCAACCCCGAAGAAAAGAGCGCTTCTCACACTAAATTCTTCTTCTCC
AAGCTTAGATTCTGA>TTHERM_00780960(gene)
TGATAATTGCTAGTTTTGGGAGAGGGTATAAATTAAAAAGCGAAAATAAAAAATATGAGT
GGGAAATAGTAAATACAAAAGAAATTAAATTAATAAGGAAACAAAAAGAGATACTTATAT
ATCTTAATAAAGAAAAAAAAGAAAAAAAAACATTATTATACAAACAAAATCAAGAAGATC
AAAAATGGCCAAATTCCTCAAGTACGGTAGAGTTGTCATCCTTTTACAAGGTAGATTCGC
TGGTAAGAAGGCTGTCATCGTCAAGTCCTCTGAAGATGGTACCAAGGACAGAAAGTTCGG
TCACGTCTTAGTTGCTGGTGTTGAAAGATCCCCCAAGAAGGTCACCAAGAGAATGGGTTC
CAAGAAGATCTAAAAGAGAACTTCCGTCAAGCCCTTCATTAAATACGTCAACCTTAACCA
TATCATGCCCACCAGATACTCCGTTAAGGAACTCTGTGACTTCAAGGAACTTGTTAAGGA
AGATAAGATCAAGAACAACGCTAAGTCTGAAGTTAGAGACACCTTAAAGAAGGTCTTCGT
CGAAAAGTACAGAACCATCAACCCCGAAGAAAAGAGCGCTTCTCACACTAAATTCTTCTT
CTCCAAGCTTAGATTCTGAGCAATTAAAAATTTTAATAACTATCCAAATATACTATAACA
ATAAATTTAATGTGTACTCATATAAAATAATCGGTTAGTATAATCCTATGAATGCTACAT
>TTHERM_00780960(protein)
MAKFLKYGRVVILLQGRFAGKKAVIVKSSEDGTKDRKFGHVLVAGVERSPKKVTKRMGSK
KIQKRTSVKPFIKYVNLNHIMPTRYSVKELCDFKELVKEDKIKNNAKSEVRDTLKKVFVE
KYRTINPEEKSASHTKFFFSKLRF