Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00823980Standard Name
PARP4 (Poly [ADP-Ribose] Polymerase 4)Aliases
PreTt20466 | 141.m00123 | 3682.m00057Description
PARP4 poly(ADP-ribose) polymerase family WGR domain protein; WGR domain containing protein; ADP-ribosyltransferase diphtheria toxin-likeGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- CURI complex (IEA) | GO:0032545
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- external side of plasma membrane (IEA) | GO:0009897
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- obsolete heterogeneous nuclear ribonucleoprotein complex (IEA) | GO:0030530
- obsolete small cytoplasmic ribonucleoprotein complex (IEA) | GO:0030531
- peptidase complex (IEA) | GO:1905368
- RQC complex (IEA) | GO:1990112
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- axon guidance receptor activity (IEA) | GO:0008046
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- carboxyvinyl-carboxyphosphonate phosphorylmutase activity (IEA) | GO:0008807
- catechol O-methyltransferase activity (IEA) | GO:0016206
- dAMP binding (IEA) | GO:0032562
- glutamate-putrescine ligase activity (IEA) | GO:0034024
- microtubule severing ATPase activity (IEA) | GO:0008568
- mitochondrial single-subunit type RNA polymerase binding (IEA) | GO:0001001
- obsolete exogenous peptide receptor activity (IEA) | GO:0008530
- obsolete gp130 (IEA) | GO:0004898
- obsolete insulin (IEA) | GO:0016088
- obsolete proprotein convertase 2 activator activity (IEA) | GO:0016484
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- proteinase activated receptor binding (IEA) | GO:0031871
- R-linalool synthase activity (IEA) | GO:0034008
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- uroporphyrinogen decarboxylase activity (IEA) | GO:0004853
- xanthine dehydrogenase activity (IEA) | GO:0004854
Biological Process
- aminoacyl-tRNA metabolism involved in translational fidelity (IEA) | GO:0106074
- basement membrane organization (IEA) | GO:0071711
- callus formation (IEA) | GO:1990110
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- endoderm development (IEA) | GO:0007492
- fibrous ring of heart morphogenesis (IEA) | GO:1905285
- garland cell differentiation (IEA) | GO:0007514
- general transcription from RNA polymerase II promoter (IEA) | GO:0032568
- haltere development (IEA) | GO:0007482
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- imaginal disc-derived genitalia development (IEA) | GO:0007484
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- laminaritriose transport (IEA) | GO:2001097
- mitochondrial fusion (IEA) | GO:0008053
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of interleukin-1 beta production (IEA) | GO:0032691
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of MyD88-independent toll-like receptor signaling pathway (IEA) | GO:0034128
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of renal water transport (IEA) | GO:2001152
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- notum development (IEA) | GO:0007477
- obsolete actin filament bundle convergence involved in mitotic contractile ring assembly (IEA) | GO:1903478
- obsolete lymph gland development (IEA) | GO:0007515
- obsolete negative regulation by organism of symbiont T-cell mediated immune response (IEA) | GO:0052474
- obsolete prothoracic morphogenesis (IEA) | GO:0007471
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- parathion metabolic process (IEA) | GO:0018952
- phagocytosis (IEA) | GO:0006909
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antifungal innate immune response (IEA) | GO:1905036
- positive regulation of dipeptide transmembrane transport (IEA) | GO:2001150
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of mating projection assembly (IEA) | GO:1902917
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of renal water transport (IEA) | GO:2001153
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- prenol metabolic process (IEA) | GO:0016090
- priming of cellular response to stress (IEA) | GO:0080136
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of D-xylose catabolic process (IEA) | GO:0043469
- regulation of dipeptide transmembrane transport (IEA) | GO:2001148
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of L-arginine import into cell (IEA) | GO:1902826
- regulation of membrane lipid metabolic process (IEA) | GO:1905038
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of respiratory gaseous exchange by nervous system process (IEA) | GO:0002087
- regulation of TOR signaling (IEA) | GO:0032006
- regulation of TRAIL production (IEA) | GO:0032679
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- vesicle coating (IEA) | GO:0006901
- visceral mesoderm-endoderm interaction involved in midgut development (IEA) | GO:0007495
- wing disc morphogenesis (IEA) | GO:0007472
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
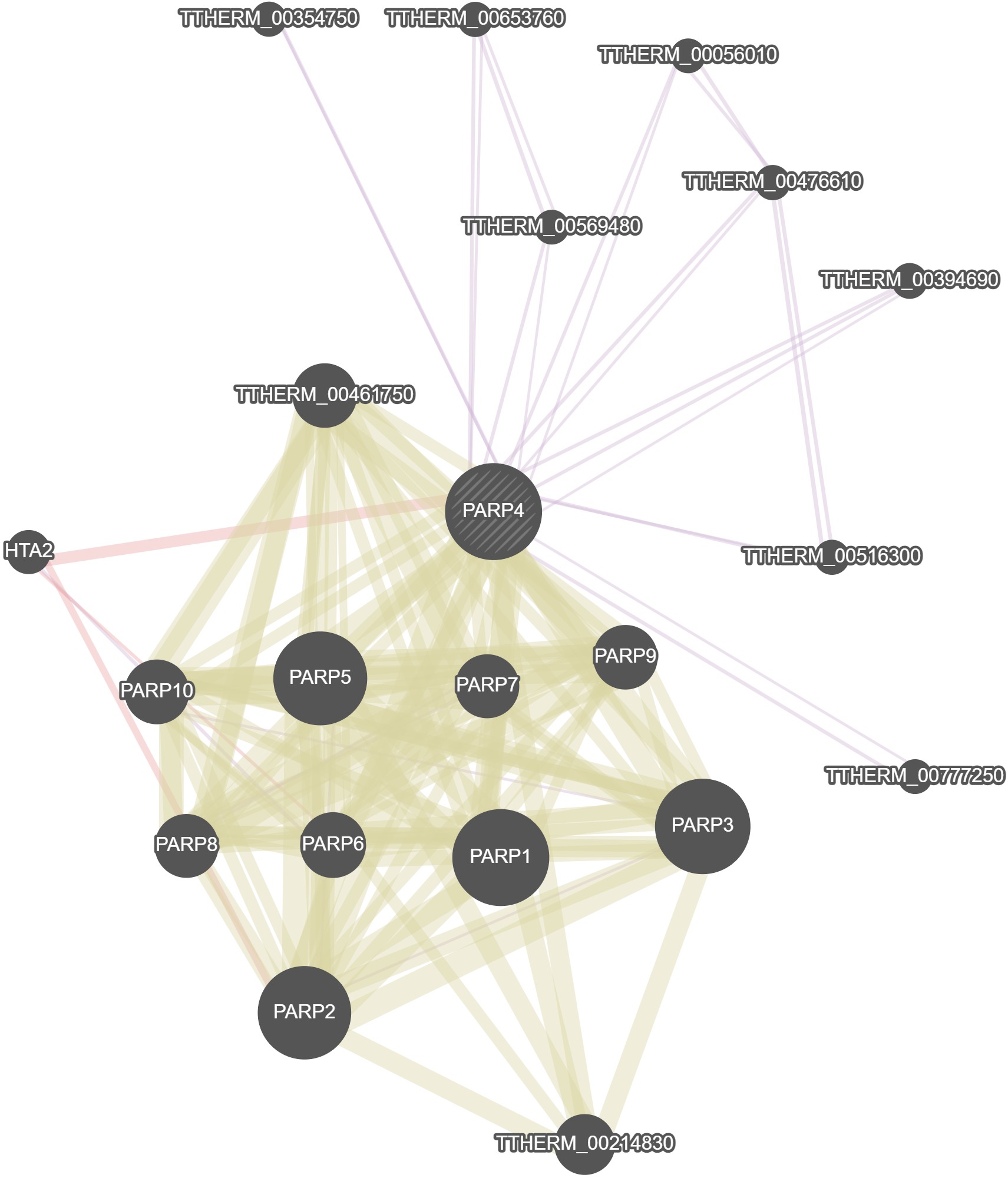
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:30796450: Ashraf K, Nabeel-Shah S, Garg J, Saettone A, Derynck J, Gingras AC, Lambert JP, Pearlman RE, Fillingham J (2019) Proteomic analysis of histones H2A/H2B and variant Hv1 in Tetrahymena thermophila reveals an ancient network of chaperones. Molecular biology and evolution ( ):
 Sequences
Sequences
>TTHERM_00823980(coding)
ATGCCTCCTAAAAGAGCTACTAGATCTTAATCTGCAGCAGCAGCAACTAAAGGTAAATAG
GCTGCTACAAAAACTACAAAAAAAGATGCCAAGATTGAGGATAAAAAAGTGACTAGAGGA
AGAAAAGCAGCCACTAAATCTCCTTCTCCTTCCCCTTCACCTAAAAAAACCCAAAAAGGT
AAACCTGCAGCAGCTTCTAAATCTAAATCTAAAGATACTAAAGTCGCAACAGTAGATAAG
AAAAACACAAAAGCAGCTTAGAAAACTACTTAAAAAACAAAGTAAGCAAAAACAGCACCA
GCACCTTCTCCCAAAAAAACCAATAAAAGAAAAGCTAAGAGTGTATCTCCCTCCCCAGAT
ACAAAGGCCTAAACTAAAAAGAGTCCTGTCAAAAAAGTCAAAGATGTACCCAAAAAAGAT
GAATCTGAAGATGAGAAAAAAACTGTTAAATAGATCGTTAAAGGTAAATGTGCAGTTGAT
TAGTATGTACCAAATTCAACACAATACACAGTTCTATAAGAAGGAAACTAAATTTACAAT
CTTACTTTGAATCAATCAAATGTTGATGCAAACAATAACAAATTCTATCTTGGATAGGTG
CTTGTTAAAGATGGAACTAATCAATACTCCGTTTTCTTTAGATGGGGAAGAGTTGGTGTT
CCAGGACAATAAAGCATTAAAGTTTGTTCAAGTAAATTTGAAGCTATTAGCGAATTCTAA
AAAAAGAAAAGCGATAAAATTAAAGGAGGTTATACTGAAATTTTCATCAAATATGGTGAA
GATGATGAAGATGAAAAGCCAAAGAAAAAATAAACTACTTAACCTACAAAGAGAAAAGAT
TGTACTTTATCTGCTGCAGTTCAAACCTTGATAAACGACATTTTTGATATGAAAATGATT
GAGAGTACAATTAAAGAAATAGGTTATGATTCTAAAAAGATGCCTTTAGGTAAACTTGAC
GATAAAACAATTAAATAAGCTTACACTGTTCTCAAAGATCTTTTAGATGCTGTAAAAAAT
AATAAAGTTAGTGATTATGGTCGTTATTCTAGTTAATTCTACTCCCTTATTCCTCATGAT
TTTGGTTTCTAAAAAATGAGTAACTTCATTTTAAAAACTGAGGAATAAGTGCGTAAGAAG
CTTTCAATGCTTTAAGATTTATCTGACTTAAAAATTGCAACTCAATTACTTAAAGAATCA
GATTCAGATGACTCAATCTTAGATTAAAACTATAAAAAATTAAATGCAGAAATAAAAGAA
GTCACAGATAAAAAGACATTAGACATTATTAAGAATTATGTTAATCAAGGCAAAGGACAT
TATAGCCCTAAGATTTTAGAAATATTTTCAGTAAAAAGAAAAGGCGAGCAAGAACGTTAC
TCAAAAAATATTGGAAACGATACACTCTTATGGCATGGGTCCCGTATTTCAAACTTTGTT
GGTATATTATCTTAAGGTCTTCGTATTGCTCCTCCTGAGGCTCCTGTTACTGGTTATAAT
TATGGTAAGGGTATTTACCTTGCAGATTAATTTACTAAATCTTGTGATTATTGTGCAGGT
AATAGTGATGGTATCCATTACATCATGCTCATAAAGGCTGCTCTAGGAACTCCTAACAAA
ATAGAAAAAACTGATTATAATGCTAATAATCTTCCTAAAGGAACTCATTCTTGCTGGGGT
TGGGGTACTCATGGTCCTGAAGAATTCATAACATTTAATGGAGTTAAAGTACCTAAAGGC
TAGGAAGTTAGAACTAAATCTAAGCACTATATGAAATATAACGAGTTTATCATTTATGAT
ATTGCTTAAGCTCAAATAGATTATCTAATTAGATTTAAAAAGTACTGA>TTHERM_00823980(gene)
ACAATTAAAAGTTAAATAAATTATATTAATCATTAAACATATCTAAGAATTTTTAATAAT
AAAAAGGATAAATTATATCTATTAATGCCTCCTAAAAGAGCTACTAGATCTTAATCTGCA
GCAGCAGCAACTAAAGGTAAATAGGCTGCTACAAAAACTACAAAAAAAGATGCCAAGATT
GAGGATAAAAAAGTGACTAGAGGAAGAAAAGCAGCCACTAAATCTCCTTCTCCTTCCCCT
TCACCTAAAAAAACCCAAAAAGGTAAACCTGCAGCAGCTTCTAAATCTAAATCTAAAGAT
ACTAAAGTCGCAACAGTAGATAAGAAAAACACAAAAGCAGCTTAGAAAACTACTTAAAAA
ACAAAGTAAGCAAAAACAGCACCAGCACCTTCTCCCAAAAAAACCAATAAAAGAAAAGCT
AAGAGTGTATCTCCCTCCCCAGATACAAAGGCCTAAACTAAAAAGAGTCCTGTCAAAAAA
GTCAAAGATGTACCCAAAAAAGATGAATCTGAAGATGAGAAAAAAACTGTTAAATAGATC
GTTAAAGGTAAATGTGCAGTTGATTAGTATGTACCAAATTCAACACAATACACAGTTCTA
TAAGAAGGAAACTAAATTTACAATCTTACTTTGAATCAATCAAATGTTGATGCAAACAAT
AACAAATTCTATCTTGGATAGGTGCTTGTTAAAGATGGAACTAATCAATACTCCGTTTTC
TTTAGATGGGGAAGAGTTGGTGTTCCAGGACAATAAAGCATTAAAGTTTGTTCAAGTAAA
TTTGAAGCTATTAGCGAATTCTAAAAAAAGAAAAGCGATAAAATTAAAGGAGGTTATACT
GAAATTTTCATCAAATATGGTGAAGATGATGAAGATGAAAAGCCAAAGAAAAAATAAACT
ACTTAACCTACAAAGAGAAAAGATTGTACTTTATCTGCTGCAGTTCAAACCTTGATAAAC
GACATTTTTGATATGAAAATGATTGAGAGTACAATTAAAGAAATAGGTTATGATTCTAAA
AAGATGCCTTTAGGTAAACTTGACGATAAAACAATTAAATAAGCTTACACTGTTCTCAAA
GATCTTTTAGATGCTGTAAAAAATAATAAAGTTAGTGATTATGGTCGTTATTCTAGTTAA
TTCTACTCCCTTATTCCTCATGATTTTGGTTTCTAAAAAATGAGTAACTTCATTTTAAAA
ACTGAGGAATAAGTGCGTAAGAAGCTTTCAATGCTTTAAGATTTATCTGACTTAAAAATT
GCAACTCAATTACTTAAAGAATCAGATTCAGATGACTCAATCTTAGATTAAAACTATAAA
AAATTAAATGCAGAAATAAAAGAAGTCACAGATAAAAAGACATTAGACATTATTAAGAAT
TATGTTAATCAAGGCAAAGGACATTATAGCCCTAAGATTTTAGAAATATTTTCAGTAAAA
AGAAAAGGCGAGCAAGAACGTTACTCAAAAAATATTGGAAACGATACACTCTTATGGCAT
GGGTCCCGTATTTCAAACTTTGTTGGTATATTATCTTAAGGTCTTCGTATTGCTCCTCCT
GAGGCTCCTGTTACTGGTTATAATTATGGTAAGGGTATTTACCTTGCAGATTAATTTACT
AAATCTTGTGATTATTGTGCAGGTAATAGTGATGGTATCCATTACATCATGCTCATAAAG
GCTGCTCTAGGAACTCCTAACAAAATAGAAAAAACTGATTATAATGCTAATAATCTTCCT
AAAGGAACTCATTCTTGCTGGGGTTGGGGTACTCATGGTCCTGAAGAATTCATAACATTT
AATGGAGTTAAAGTACCTAAAGGCTAGGAAGTTAGAACTAAATCTAAGCACTATATGAAA
TATAACGAGTTTATCATTTATGATATTGCTTAAGCTCAAATAGATTATCTAATTAGATTT
AAAAAGTACTGAGTTAGTTTAAAGTTATTATAAGCCATATTTCTTATGAAAATTTATTTG
ATAGTTTGTTTGAATATTTATTTTATGCCTTGTATTGTATTTTTATTGAAATAAATTAAT
TGGTAACTTGATTTTTATTAAATTTTTTTTTAAAAT>TTHERM_00823980(protein)
MPPKRATRSQSAAAATKGKQAATKTTKKDAKIEDKKVTRGRKAATKSPSPSPSPKKTQKG
KPAAASKSKSKDTKVATVDKKNTKAAQKTTQKTKQAKTAPAPSPKKTNKRKAKSVSPSPD
TKAQTKKSPVKKVKDVPKKDESEDEKKTVKQIVKGKCAVDQYVPNSTQYTVLQEGNQIYN
LTLNQSNVDANNNKFYLGQVLVKDGTNQYSVFFRWGRVGVPGQQSIKVCSSKFEAISEFQ
KKKSDKIKGGYTEIFIKYGEDDEDEKPKKKQTTQPTKRKDCTLSAAVQTLINDIFDMKMI
ESTIKEIGYDSKKMPLGKLDDKTIKQAYTVLKDLLDAVKNNKVSDYGRYSSQFYSLIPHD
FGFQKMSNFILKTEEQVRKKLSMLQDLSDLKIATQLLKESDSDDSILDQNYKKLNAEIKE
VTDKKTLDIIKNYVNQGKGHYSPKILEIFSVKRKGEQERYSKNIGNDTLLWHGSRISNFV
GILSQGLRIAPPEAPVTGYNYGKGIYLADQFTKSCDYCAGNSDGIHYIMLIKAALGTPNK
IEKTDYNANNLPKGTHSCWGWGTHGPEEFITFNGVKVPKGQEVRTKSKHYMKYNEFIIYD
IAQAQIDYLIRFKKY