Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00865250Standard Name
PPI8 (peptidylprolyl isomerase 8)Aliases
PreTt29155 | 154.m00113 | 3751.m00459Description
PPI8 peptidyl-prolyl cis-trans isomerase; PPIC-type PPIASE domain containing protein; Peptidyl-prolyl cis-trans isomerase Pin1Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
No Data fetched for Gene Ontology Annotations
 Domains
Domains
- ( PF00639 ) PPIC-type PPIASE domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
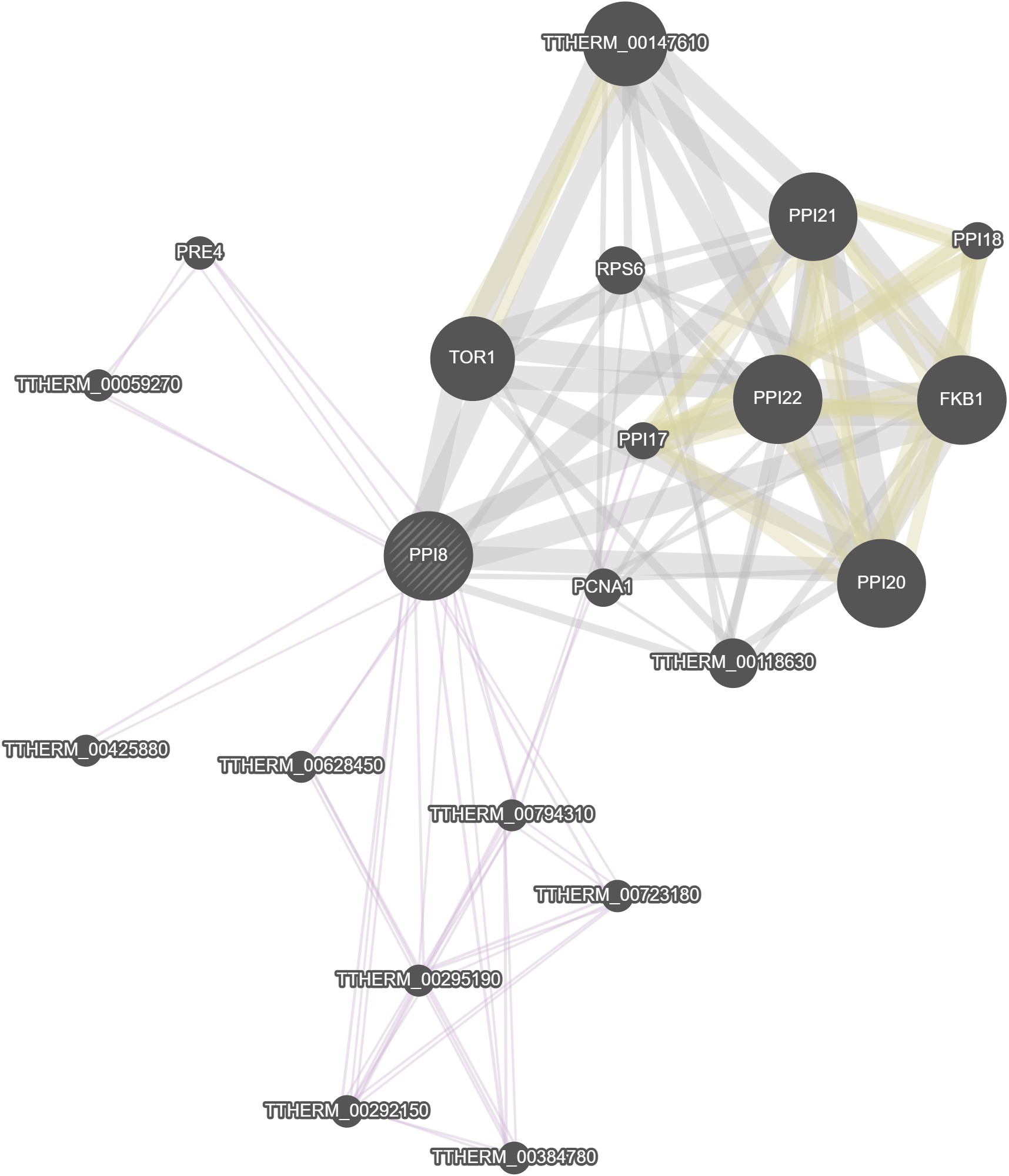
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00865250(coding)
ATGAACTCTTAAACAATCAGAGCTGCTCATATTTTATAAAAGCATAGAGGAAGTCGTAAT
CCACTTGATAGAGTTAGAAATGTCCAAGTTACTAGAACTTTAGATGAAGCTAAAAAAAAT
GTAGCAGCTTTCCGTGAATAGATCATGAAATCAGCTGATCCTTAAAAAACTTTTATGGAA
ATTGCACAAAAGTATTCTGAATGCACATCTGCTAGAAATGGAGGTGACTTGGGAGAGTTT
GGTCCTGGTTAAATGCAAGAATCATTTGAATAGGCTGCATATGCTCTTAAAGTCGGAGAG
ATTTCTAACTTAGTTGAGTCAGACTCTGGTGTTCACATTATTTTAAGATTAGATTGA>TTHERM_00865250(gene)
AAAAAAAAATATTTTTTATTCAATTTATTATTAAAGTAAAAATAAAATAAAATATGAATT
TTTGAAAGTATCCGTTGTTGTTGGCTAAGATTTGTTGCTTTTATTTTTAAGATCAGAAAG
TTACAAGAATTAATATTCAAGTAGAGAAAATTTGAAATTAAAAATAAATTAGTCACTGAA
TTAAAAAAAAATAAATAATCTCTTAAAAATCACTAAATTTAATATTAAAAAAAAGCAAAT
AATCATAAATGAACTCTTAAACAATCAGAGCTGCTCATATTTTATAAAAGCATAGAGGAA
GTCGTAATCCACTTGATAGAGTTAGAAATGTCCAAGTTACTAGAACTTTAGATGAAGCTA
AAAAAAATGTAGCAGCTTTCCGTGAATAGATCATGAAATCAGCTGATCCTTAAAAAACTT
TTATGGAAATTGCACAAAAGTATTCTGAATGCACATCTGCTAGAAATGGAGGTGACTTGG
GAGAGTTTGGTCCTGGTTAAATGCAAGAATCATTTGAATAGGCTGCATATGCTCTTAAAG
TCGGAGAGATTTCTAACTTAGTTGAGTCAGACTCTGGTGTTCACATTATTTTAAGATTAG
ATTGATTTAAAAGTAAAATGTGATAAAATAAACAAATAAAAATAATATGAATCTGATTGA
TAAAAGAACATAATAATTAATAATATAAATACTTTAAAATTCATATTTTTATAATTAGTT
GCATCATCCTTAAACAGTTTAATTGTTTATTCAAAA>TTHERM_00865250(protein)
MNSQTIRAAHILQKHRGSRNPLDRVRNVQVTRTLDEAKKNVAAFREQIMKSADPQKTFME
IAQKYSECTSARNGGDLGEFGPGQMQESFEQAAYALKVGEISNLVESDSGVHIILRLD