Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00957580Standard Name
RAB17A (RAB GTPase 17A)Aliases
PreTt21654 | 177.m00058Description
RAB17A small guanosine triphosphatase family Ras family protein; Rab V family protein; small monomeric GTPase that regulates membrane fusion and fission events; Small GTPaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- small-subunit processome (IEA) | GO:0032040
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 2'-phosphotransferase activity (IEA) | GO:0008665
- 2',3'-cyclic-nucleotide 2'-phosphodiesterase activity (IEA) | GO:0008663
- ADP phosphatase activity (IEA) | GO:0043262
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- insulin-like growth factor II binding (IEA) | GO:0031995
- ion channel inhibitor activity (IEA) | GO:0008200
- kappa-type opioid receptor binding (IEA) | GO:0031851
- monoatomic anion transmembrane transporter activity (IEA) | GO:0008509
- obsolete endogenous peptide receptor activity (IEA) | GO:0008529
- obsolete phospholipid scrambling (IEA) | GO:0008498
- obsolete serpin (IEA) | GO:0004868
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylinositol-3-phosphate binding (IEA) | GO:0032266
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- replication fork barrier binding (IEA) | GO:0031634
- RNA 2',3'-cyclic 3'-phosphodiesterase activity (IEA) | GO:0008664
- sodium:potassium:chloride symporter activity (IEA) | GO:0008511
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sucrose:proton symporter activity (IEA) | GO:0008506
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- allantoate transport (IEA) | GO:0015719
- bioluminescence (IEA) | GO:0008218
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- central nervous system myelin maintenance (IEA) | GO:0032286
- commissural neuron axon guidance (IEA) | GO:0071679
- embryonic brain development (IEA) | GO:1990403
- glycolytic process through glucose-1-phosphate (IEA) | GO:0061622
- glycolytic process through glucose-6-phosphate (IEA) | GO:0061620
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-19 production (IEA) | GO:0032622
- interleukin-2 production (IEA) | GO:0032623
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- mitotic nuclear division (IEA) | GO:0007067
- modulation by organism of innate immune response in other organism involved in symbiotic interaction (IEA) | GO:0052306
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of glycine secretion, neurotransmission (IEA) | GO:1904625
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phospholipid translocation (IEA) | GO:0061093
- nucleotide-binding oligomerization domain containing 2 signaling pathway (IEA) | GO:0070431
- obsolete chemi-mechanical coupling (IEA) | GO:0006943
- obsolete histone H3-K4 trimethylation (IEA) | GO:0080182
- obsolete negative regulation of cGMP catabolic process (IEA) | GO:0030830
- obsolete positive regulation of transcription from RNA polymerase II promoter in response to maltose (IEA) | GO:1904765
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- phenanthrene catabolic process via trans-9(R),10(R)-dihydrodiolphenanthrene (IEA) | GO:0018956
- photosynthetic NADP+ reduction (IEA) | GO:0009780
- plant-type cell wall modification (IEA) | GO:0009827
- polyamine acetylation (IEA) | GO:0032917
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of basidium development (IEA) | GO:0075315
- positive regulation of fatty acid beta-oxidation (IEA) | GO:0032000
- positive regulation of fibroblast growth factor receptor signaling pathway involved in ureteric bud formation (IEA) | GO:2000704
- pre-replicative complex assembly involved in cell cycle DNA replication (IEA) | GO:1902299
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- prostate induction (IEA) | GO:0060514
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of nucleotide-excision repair (IEA) | GO:2000819
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- regulation of striated muscle contraction (IEA) | GO:0006942
- response to photooxidative stress (IEA) | GO:0080183
- single strand break repair (IEA) | GO:0000012
- striated muscle contraction (IEA) | GO:0006941
 Domains
Domains
- ( PF00071 ) Ras family
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
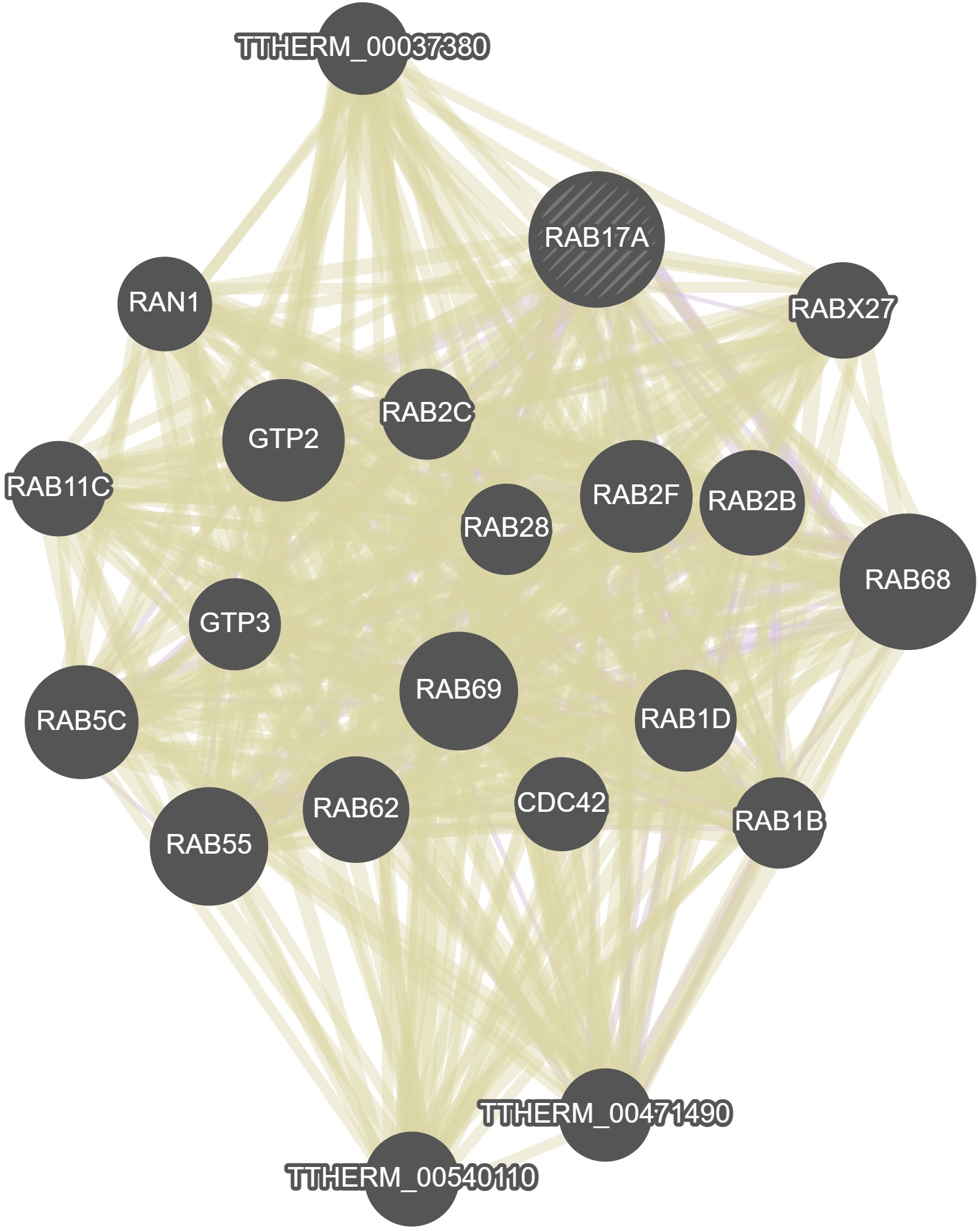
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_00957580(coding)
ATGAAGCATAAGTATATCTAATAAAATATAAAGACAAGTTTATCAAAAAAAAAAAAAAAT
TAAATAATAGGAAAAACAAACTATAAAATTTTGCAAATAAATATCCTTAATTAAAAATCA
TATCAAATTTGCATTCAAATAATGTGTTTCGGATCCAGCTCTAAGAAATAGTAAAGCAAA
CCCAAGCTTTTAAAGCCCAAGCAAAGTTAGTACAAAACTAAGCTCATTCTTTTAGGTGAC
ATGAACGTAGGCAAAACTTCAATCTGCGAGTATTACAAGAAGAACGTGCCTGCAAAGAAC
TTGCCAAATACAATTGGTGTCACATATTTCGAGTTCGACATCAAACTAGAAAATGGCATT
AACGTTCACTTAGATATTTGGGATACAGCAGGATAAGAAATTTATTAGTCACTACTAAAG
ATATACTATCAAGATGCTAACATAGTTTTTTTAGTTTTTGATGTGAGTGCAAGATAGACT
CTTGAACGTGTTGAGTACTGGATTAGCGAACTCAAAGAGCAAGTAGATGATAAAAACCTA
AAGTTGGTGTTAATAGGTAACAAATGCGACATAGATGCAGATAAGAGGTAAATCAGTCTT
GAGCAAGGTCAAGATTTTGCTAGAAATAAAAACATTCCTTACTTTGAAACTAGTGCAATC
ACTGGAGAAGGAATAGTTGAAGTATTCCGCTAAACAGTAAAGGACTATGTGTAATATAAG
ACAAACGGAGGAAAATGA>TTHERM_00957580(gene)
TTTTTTGTTTATTGCTGGTTTGATATGAAGCATAAGTATATCTAATAAAATATAAAGACA
AGTTTATCAAAAAAAAAAAAAAATTAAATAATAGGAAAAACAAACTATAAAATTTTGCAA
ATAAATATCCTTAATTAAAAATCATATCAAATTTGCATTCAAATAATGTGTTTCGGATCC
AGCTCTAAGAAATAGTAAAGCAAACCCAAGCTTTTAAAGCCCAAGCAAAGTTAGTACAAA
ACTAAGCTCATTCTTTTAGGTGACATGAACGTAGGCAAAACTTCAATCTGCGAGTATTAC
AAGAAGAACGTGCCTGCAAAGAACTTGCCAAATACAATTGGTGTCACATATTTCGAGTTC
GACATCAAACTAGAAAATGGCATTAACGTTCACTTAGATATTTGGGATACAGCAGGATAA
GAAATTTATTAGTCACTACTAAAGATATACTATCAAGATGCTAACATAGTTTTTTTAGTT
TTTGATGTGAGTGCAAGATAGACTCTTGAACGTGTTGAGTACTGGATTAGCGAACTCAAA
GAGCAAGTAGATGATAAAAACCTAAAGTTGGTGTTAATAGGTAACAAATGCGACATAGAT
GCAGATAAGAGGTAAATCAGTCTTGAGCAAGGTCAAGATTTTGCTAGAAATAAAAACATT
CCTTACTTTGAAACTAGTGCAATCACTGGAGAAGGAATAGTTGAAGTATTCCGCTAAACA
GTAAAGGACTATGTGTAATATAAGACAAACGGAGGAAAATGAATATAACAATTAATTAAT
CACTCATAAATAAAATTAAATCCTCAAAGCAAATGAATAATGCCATTTGCTTTTGGCATA
ATCAATTTCTCTTCTTTATTTCTTTCTTTCTTTCTAAGACTGACATAGTAAAATAAATAA
ACAAATAAATATATTTAAGCTACGACTAGATAAAAATTAATAAATGTAAAAATAATTAGA
AAAATAGAATAGATTTATAAATAATAAAATAGCTTATAAGTAAATAAATAAATAAATAAA
TAAAAAAACAATAAATCAACTAAACAACAACAATAACAACCAACCAACCAAAATAACTTT
TAAAATCTTTAAATTTATTTATCCTTTAATAAAAAATAATAAAAACAATTAAACAAATGT
GAATTAAAATTGAATGAATAAATGAATGAATGATTTCTTTAGTCATATTTAACTTTATAT
TCTACTTGTTCTTCTTTTATAAAACAAATACTATTCTCAA>TTHERM_00957580(protein)
MKHKYIQQNIKTSLSKKKKNQIIGKTNYKILQINILNQKSYQICIQIMCFGSSSKKQQSK
PKLLKPKQSQYKTKLILLGDMNVGKTSICEYYKKNVPAKNLPNTIGVTYFEFDIKLENGI
NVHLDIWDTAGQEIYQSLLKIYYQDANIVFLVFDVSARQTLERVEYWISELKEQVDDKNL
KLVLIGNKCDIDADKRQISLEQGQDFARNKNIPYFETSAITGEGIVEVFRQTVKDYVQYK
TNGGK