Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00999090Standard Name
SLP1 (Scramblase-Like Protein 1)Aliases
PreTt26088 | 189.m00066 | 3819.m00220Description
SLP1 scramblase family protein; Scramblase family protein; redistributes plasma membrane asymmetry by transporting negatively charged phospholipids from inner leaflet to outer leaflet of lipid bilayer; ScramblaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- cytoplasm (IDA) | GO:0005737
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- gamma-catenin-TCF7L2 complex (IEA) | GO:0071665
- glutamate dehydrogenase complex (IEA) | GO:1990148
- motile primary cilium (IEA) | GO:0031512
- plasma membrane (IDA) | GO:0005886
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- C5L2 anaphylatoxin chemotactic receptor binding (IEA) | GO:0031715
- cAMP-dependent protein kinase regulator activity (IEA) | GO:0008603
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- fucosyltransferase activity (IEA) | GO:0008417
- G protein-coupled serotonin receptor binding (IEA) | GO:0031821
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- microtubule severing ATPase activity (IEA) | GO:0008568
- obsolete bilirubin secondary active transmembrane transporter activity (IEA) | GO:0015351
- obsolete octanoyltransferase activity (IEA) | GO:0016415
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete prostaglandin/thromboxane transporter activity (IEA) | GO:0015348
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- protein kinase C inhibitor activity (IEA) | GO:0008426
- pyridoxal kinase activity (IEA) | GO:0008478
- sodium-independent organic anion transmembrane transporter activity (IEA) | GO:0015347
- sterol binding (IEA) | GO:0032934
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- allantoate transport (IEA) | GO:0015719
- apoptotic mitochondrial changes (IEA) | GO:0008637
- bile acid biosynthetic process (IEA) | GO:0006699
- bronchus development (IEA) | GO:0060433
- bronchus morphogenesis (IEA) | GO:0060434
- carbon utilization (IEA) | GO:0015976
- carotenoid metabolic process (IEA) | GO:0016116
- cytoplasmic actin-based contraction involved in forward cell motility (IEA) | GO:0060328
- cytoplasmic actin-based contraction involved in rearward cell motility (IEA) | GO:0060329
- endoplasmic reticulum membrane fusion (IEA) | GO:0016320
- energy derivation by oxidation of reduced inorganic compounds (IEA) | GO:0015975
- glycolytic process from glycerol (IEA) | GO:0061613
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- loss of chromatin silencing (IEA) | GO:0006345
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- mesonephros development (IEA) | GO:0001823
- mitotic nuclear division (IEA) | GO:0007067
- negative regulation of brain-derived neurotrophic factor receptor signaling pathway (IEA) | GO:0031549
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine-mediated signaling pathway (IEA) | GO:0070100
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of type I interferon-mediated signaling pathway (IEA) | GO:0060339
- neuron remodeling (IEA) | GO:0016322
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- phagocytosis, engulfment (IEA) | GO:0006911
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of activin receptor signaling pathway (IEA) | GO:0032927
- positive regulation of ATF6-mediated unfolded protein response (IEA) | GO:1903893
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of somitomeric trunk muscle development (IEA) | GO:0014709
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine deoxyribonucleotide biosynthetic process (IEA) | GO:0009221
- pyrimidine ribonucleotide catabolic process (IEA) | GO:0009222
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of dendrite extension (IEA) | GO:1903859
- regulation of interleukin-27-mediated signaling pathway (IEA) | GO:0070107
- regulation of protein maturation by peptide bond cleavage (IEA) | GO:0010953
- regulation of response to type II interferon (IEA) | GO:0060330
- regulation of smooth muscle cell chemotaxis (IEA) | GO:0071671
- sperm aster formation (IEA) | GO:0035044
- steroid hormone receptor complex assembly (IEA) | GO:0006463
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
 Domains
Domains
- ( PF03803 ) Scramblase
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
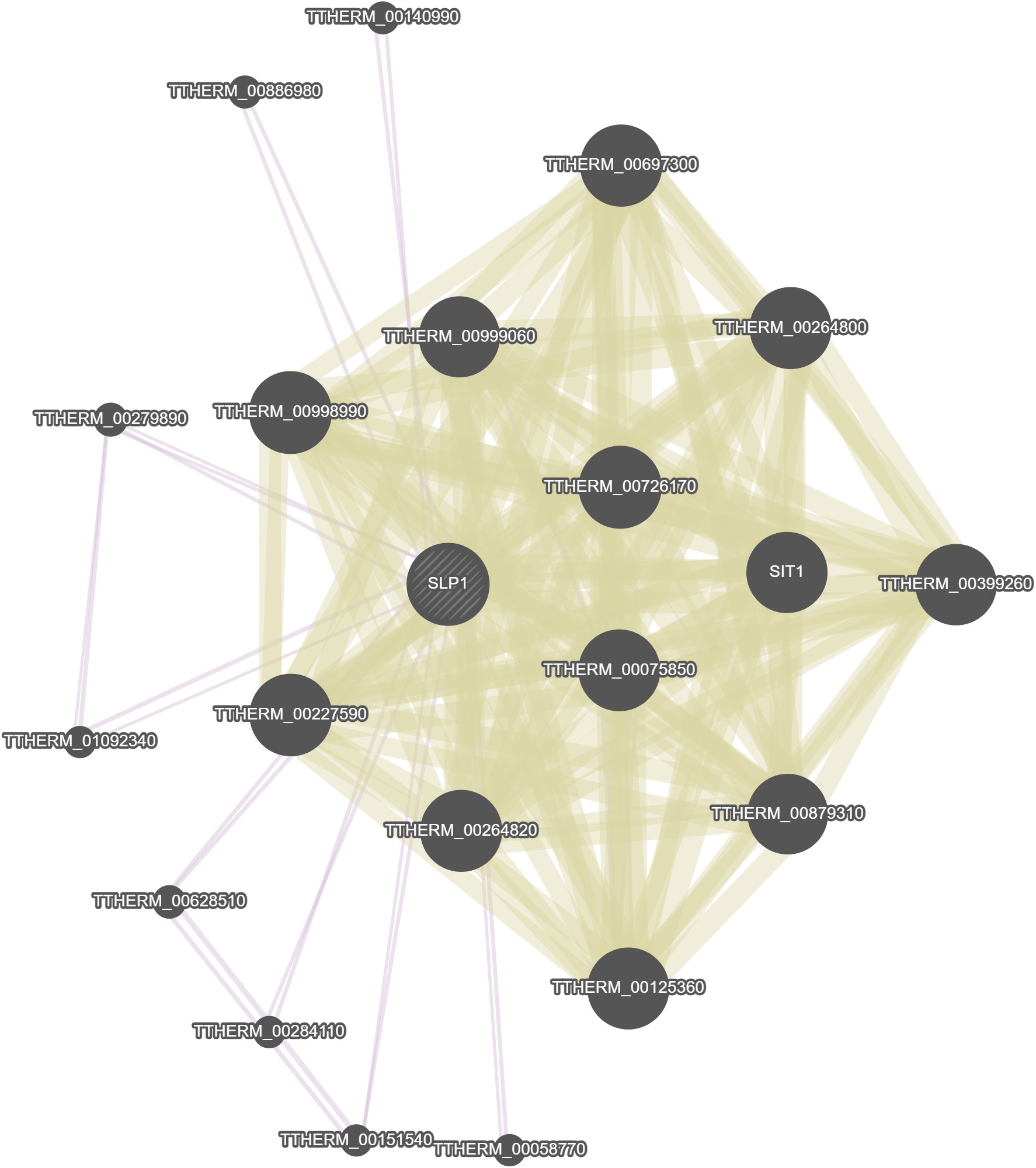
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_00999090(coding)
ATGTATCCACCATAATAGCAATAGTATAATCAATAACCCTATGGTTAAGGTTATGGATAA
TAAGGTATGCCTAACATTTAAATGATGCCTATGCAATAGTATGCAGTAGTGGCTATTTAA
AACGGATTGTAAAAGTTAGCTTCTGCTTAAGGTATATTTATTGAATAGAAGTTAGAAGTC
ATTGAGGCATTGACTGGATGCTAAACTCCTAATGTCTACAAGGTTTATCCTGCTGATGTT
AATGGTATATAAACATCAGATTAAACTATATTCAAGTGTAAAGAACTTAGTAACTGTTGT
GTACGTTAATGCATAGCCCCAAGCTGCCGTCCATTTGATATGGCAGTCACAAATTAGCAA
GGAAAATTAAATTATGGATAACTTTCTGGCTCAACTTTTATTTAAATGTAGCGTCCATTC
AAGTGTACATGCCTTTGCTTCAACCGTCCTGAACTCTCTGTAACAGTTGTAGAAAATGGG
ATGAATTAGTATCTAGGAAAAATTAGAGAACCATTTAGATTTTGTGATCTTTGCGTTGAA
ATTGAAGATGCTGCAGGCAATCTCAAGTTTATAATTTCTGGATCCTGCTGCTAACTTGGT
TATCTTTGTATGGGTTGCCCTTATCAATCTTGCCAAGAATTAACTTTTGAAGTAAAGAAT
CCAGCAGGTGAACTAATTACAAGAATTTTAAAGAAATCTGCAGGTTGTCTAAAGAGTTGT
TTTGGTAAAACAGATAACTTTACTATAGGTTTCCCTCCTGGTTTGTCTCCTGAAGACAAA
GCTCTTTTTATGGCTGCTACTCTTTTAATTGATTATATGTACTTTGAAAATAAAACAGAA
CAACCAAATGATTTATGA>TTHERM_00999090(gene)
TCTATTTATAAATAATTTTTTATTTATTTACCTATTCATATATTTATTGCGAAATATAAT
TGTTATAAATATTAATTATCATATAAATTTAATAAAAAGTTAAGTAAAATAAATTATCAT
CAATAAATAAATGTATCCACCATAATAGCAATAGTATAATCAATAACCCTATGGTTAAGG
TTATGGATAATAAGGTATGCCTAACATTTAAATGATGCCTATGCAATAGTATGCAGTAGT
GGCTATTTAAAACGGATTGTAAAAGTTAGCTTCTGCTTAAGGTATATTTATTGAATAGAA
GTTAGAAGTCATTGAGGCATTGACTGGATGCTAAACTCCTAATGTCTACAAGGTTTATCC
TGCTGATGTTAATGGTATATAAACATCAGATTAAACTATATTCAAGTGTAAAGAACTTAG
TAACTGTTGTGTACGTTAATGCATAGCCCCAAGCTGCCGTCCATTTGATATGGCAGTCAC
AAATTAGCAAGGAAAATTAAATTATGGATAACTTTCTGGCTCAACTTTTATTTAAATGTA
GCGTCCATTCAAGTGTACATGCCTTTGCTTCAACCGTCCTGAACTCTCTGTAACAGTTGT
AGAAAATGGGATGAATTAGTATCTAGGAAAAATTAGAGAACCATTTAGATTTTGTGATCT
TTGCGTTGAAATTGAAGATGCTGCAGGCAATCTCAAGTTTATAATTTCTGGATCCTGCTG
CTAACTTGGTTATCTTTGTATGGGTTGCCCTTATCAATCTTGCCAAGAATTAACTTTTGA
AGTAAAGAATCCAGCAGGTGAACTAATTACAAGAATTTTAAAGAAATCTGCAGGTTGTCT
AAAGAGTTGTTTTGGTAAAACAGATAACTTTACTATAGGTTTCCCTCCTGGTTTGTCTCC
TGAAGACAAAGCTCTTTTTATGGCTGCTACTCTTTTAATTGATTATATGTACTTTGAAAA
TAAAACAGAACAACCAAATGATTTATGAAATATCATCAAATTAAGTTATTTATTTATATA
TAAAACTTATTTAGTATATTTTTATTCATCTTACCTTTCCTATTCACATGCGTTTATGCT
AATTTCAAAGCACTTCTACCAATAAATATAATATATAAAATCTCTCACTAAAACAAACAA
ACAAACATTCAATCAATATTATATTAAAATGTTAGAACTATTAATTGATATTTTTGATGA
TATTTATTGCTTATTTTTTAAAACTAAATAAAATTTAAAGTTTATAACAA>TTHERM_00999090(protein)
MYPPQQQQYNQQPYGQGYGQQGMPNIQMMPMQQYAVVAIQNGLQKLASAQGIFIEQKLEV
IEALTGCQTPNVYKVYPADVNGIQTSDQTIFKCKELSNCCVRQCIAPSCRPFDMAVTNQQ
GKLNYGQLSGSTFIQMQRPFKCTCLCFNRPELSVTVVENGMNQYLGKIREPFRFCDLCVE
IEDAAGNLKFIISGSCCQLGYLCMGCPYQSCQELTFEVKNPAGELITRILKKSAGCLKSC
FGKTDNFTIGFPPGLSPEDKALFMAATLLIDYMYFENKTEQPNDL