Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01014460Standard Name
CST1 (Calcium-dependent Serine/Threonine kinase 1)Aliases
PreTt04576 | 197.m00050 | 3748.m00287Description
CST1 kinase domain protein; Protein kinase domain containing protein; involved in programmed nuclear death of macronucleus and reabsorption by autophagosome; Calcium-dependent Serine/Threonine Protein KinasesGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- aleurone grain membrane (IEA) | GO:0032578
- alpha6-beta1 integrin-CD151 complex (IEA) | GO:0071059
- CURI complex (IEA) | GO:0032545
- cytoplasm (IDA) | GO:0005737
- Dbf4-dependent protein kinase complex (IEA) | GO:0031431
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- endoplasmic reticulum (IEA) | GO:0005783
- multivesicular body (IEA) | GO:0005771
- vacuolar membrane (IEA) | GO:0005774
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 6-phospho-beta-galactosidase activity (IEA) | GO:0033920
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- ABC-type maltose transporter activity (IEA) | GO:0015423
- ABC-type oligosaccharide transporter activity (IEA) | GO:0015422
- acetyl-CoA transmembrane transporter activity (IEA) | GO:0008521
- aromatase activity (IEA) | GO:0008402
- arsenate reductase (glutaredoxin) activity (IEA) | GO:0008794
- axon guidance receptor activity (IEA) | GO:0008046
- carboxymethylenebutenolidase activity (IEA) | GO:0008806
- carboxyvinyl-carboxyphosphonate phosphorylmutase activity (IEA) | GO:0008807
- cardiolipin synthase activity (IEA) | GO:0008808
- cellulase activity (IEA) | GO:0008810
- Cys-tRNA(Pro) hydrolase activity (IEA) | GO:0043907
- delta5-delta2,4-dienoyl-CoA isomerase activity (IEA) | GO:0008416
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- glutamate-putrescine ligase activity (IEA) | GO:0034024
- GTPase motor activity (IEA) | GO:0061791
- insulin-like growth factor II binding (IEA) | GO:0031995
- methane monooxygenase [NAD(P)H] activity (IEA) | GO:0015049
- microtubule severing ATPase activity (IEA) | GO:0008568
- myoinhibitory hormone activity (IEA) | GO:0016085
- N-acetyl-gamma-aminoadipyl-phosphate reductase activity (IEA) | GO:0043870
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete D-lysine oxidase activity (IEA) | GO:0043912
- obsolete myosin ATPase activity (IEA) | GO:0008570
- obsolete nucleoplasmin ATPase activity (IEA) | GO:0008572
- obsolete prostaglandin/thromboxane transporter activity (IEA) | GO:0015348
- obsolete red-sensitive opsin (IEA) | GO:0015061
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete X-opioid receptor activity (IEA) | GO:0015051
- peptide alpha-N-propionyltransferase activity (IEA) | GO:0061607
- peptidoglycan stem peptide endopeptidase activity (IEA) | GO:0061786
- phosphatidylcholine:cardiolipin O-linoleoyltransferase activity (IEA) | GO:0032577
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- potassium:proton antiporter activity (IEA) | GO:0015386
- R-linalool synthase activity (IEA) | GO:0034008
- Ser(Gly)-tRNA(Ala) hydrolase activity (IEA) | GO:0043908
- sodium-independent organic anion transmembrane transporter activity (IEA) | GO:0015347
- sodium:potassium:chloride symporter activity (IEA) | GO:0008511
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sterol 14-demethylase activity (IEA) | GO:0008398
- tubulin-glutamic acid ligase activity (IEA) | GO:0070740
Biological Process
- 1-undecene metabolic process (IEA) | GO:1902820
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- acetylcholine biosynthetic process (IEA) | GO:0008292
- acetylcholine metabolic process (IEA) | GO:0008291
- antigen processing and presentation following receptor mediated endocytosis (IEA) | GO:0002751
- bronchus development (IEA) | GO:0060433
- bronchus morphogenesis (IEA) | GO:0060434
- bud dilation involved in lung branching (IEA) | GO:0060503
- cardiac endothelial cell differentiation (IEA) | GO:0003348
- cell migration in diencephalon (IEA) | GO:0061381
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to D-galactose (IEA) | GO:1905378
- cellular response to homocysteine (IEA) | GO:1905375
- cellular response to phosphate starvation (IEA) | GO:0016036
- cilium movement (IEA) | GO:0003341
- coenzyme A biosynthetic process (IEA) | GO:0015937
- dendrite development (IEA) | GO:0016358
- DNA replication preinitiation complex assembly (IEA) | GO:0071163
- endosomal pattern recognition receptor signaling pathway (IEA) | GO:0002754
- epithelial cell proliferation involved in lung bud dilation (IEA) | GO:0060505
- epithelium development (IEA) | GO:0060429
- ethyl acetate biosynthetic process (IEA) | GO:1902819
- female courtship behavior (IEA) | GO:0008050
- germinal center formation (IEA) | GO:0002467
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immune response-activating signaling pathway (IEA) | GO:0002757
- inferior colliculus development (IEA) | GO:0061379
- interleukin-12-mediated signaling pathway (IEA) | GO:0035722
- interleukin-15-mediated signaling pathway (IEA) | GO:0035723
- interleukin-28A secretion (IEA) | GO:0072628
- intraciliary retrograde transport (IEA) | GO:0035721
- laminaritriose transport (IEA) | GO:2001097
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- mesonephric podocyte cell fate commitment (IEA) | GO:0061258
- microtubule cytoskeleton organization involved in mitosis (IEA) | GO:1902850
- mitochondrial membrane organization (IEA) | GO:0007006
- multicellular organism development (IEA) | GO:0007275
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of autophagosome assembly (IEA) | GO:1902902
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of cytoplasmic translation (IEA) | GO:2000766
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of encysted zoospore germination (IEA) | GO:0075229
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of hindgut contraction (IEA) | GO:0060451
- negative regulation of immunological synapse formation (IEA) | GO:2000521
- negative regulation of peripheral B cell deletion (IEA) | GO:0002909
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of supramolecular fiber organization (IEA) | GO:1902904
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete activation of MAPKK activity (IEA) | GO:0000186
- obsolete actomyosin contractile ring disassembly (IEA) | GO:1902621
- obsolete fertilization, exchange of chromosomal proteins (IEA) | GO:0035042
- obsolete histone H4-K20 demethylation (IEA) | GO:0035574
- obsolete MAPK import into nucleus involved in osmosensory signaling pathway (IEA) | GO:0000208
- obsolete negative regulation of response to extracellular stimulus (IEA) | GO:0032105
- obsolete positive regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process (IEA) | GO:1902964
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response (IEA) | GO:0006990
- obsolete regulation of glucose-6-phosphatase activity (IEA) | GO:0032114
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- P granule disassembly (IEA) | GO:1903864
- peripheral B cell positive selection (IEA) | GO:0002350
- peripheral tolerance induction (IEA) | GO:0002465
- phagocytosis (IEA) | GO:0006909
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of DNA amplification (IEA) | GO:1904525
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of intestinal epithelial cell development (IEA) | GO:1905300
- positive regulation of late endosome to lysosome transport (IEA) | GO:1902824
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of protein autoubiquitination (IEA) | GO:1902499
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of siRNA processing (IEA) | GO:1903705
- positive regulation of sodium ion import across plasma membrane (IEA) | GO:1903784
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- priming of cellular response to stress (IEA) | GO:0080136
- proepicardium development (IEA) | GO:0003342
- programmed cell death (IDA) | GO:0012501
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- quercetin biosynthetic process (IEA) | GO:1901734
- quercetin metabolic process (IEA) | GO:1901732
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antibacterial peptide production (IEA) | GO:0002786
- regulation of cardioblast proliferation (IEA) | GO:0003264
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of gastric acid secretion (IEA) | GO:0060453
- regulation of L-arginine import into cell (IEA) | GO:1902826
- regulation of late endosome to lysosome transport (IEA) | GO:1902822
- regulation of macropinocytosis (IEA) | GO:1905301
- regulation of microtubule binding (IEA) | GO:1904526
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of secondary heart field cardioblast proliferation (IEA) | GO:0003266
- regulation of supramolecular fiber organization (IEA) | GO:1902903
- release of sequestered calcium ion into cytosol by endoplasmic reticulum (IEA) | GO:1903514
- response to transition metal nanoparticle (IEA) | GO:1990267
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- sphinganine metabolic process (IEA) | GO:0006667
- sterol regulatory element binding protein cleavage involved in ER-nuclear sterol response pathway (IEA) | GO:0006992
- synaptic vesicle docking (IEA) | GO:0016081
- synaptic vesicle priming (IEA) | GO:0016082
- T cell inhibitory signaling pathway (IEA) | GO:0002770
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- tetrasaccharide transport (IEA) | GO:2001098
- triterpenoid catabolic process (IEA) | GO:0016105
- vacuolar acidification (IEA) | GO:0007035
- xanthophyll biosynthetic process (IEA) | GO:0016123
- xanthophyll catabolic process (IEA) | GO:0016124
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
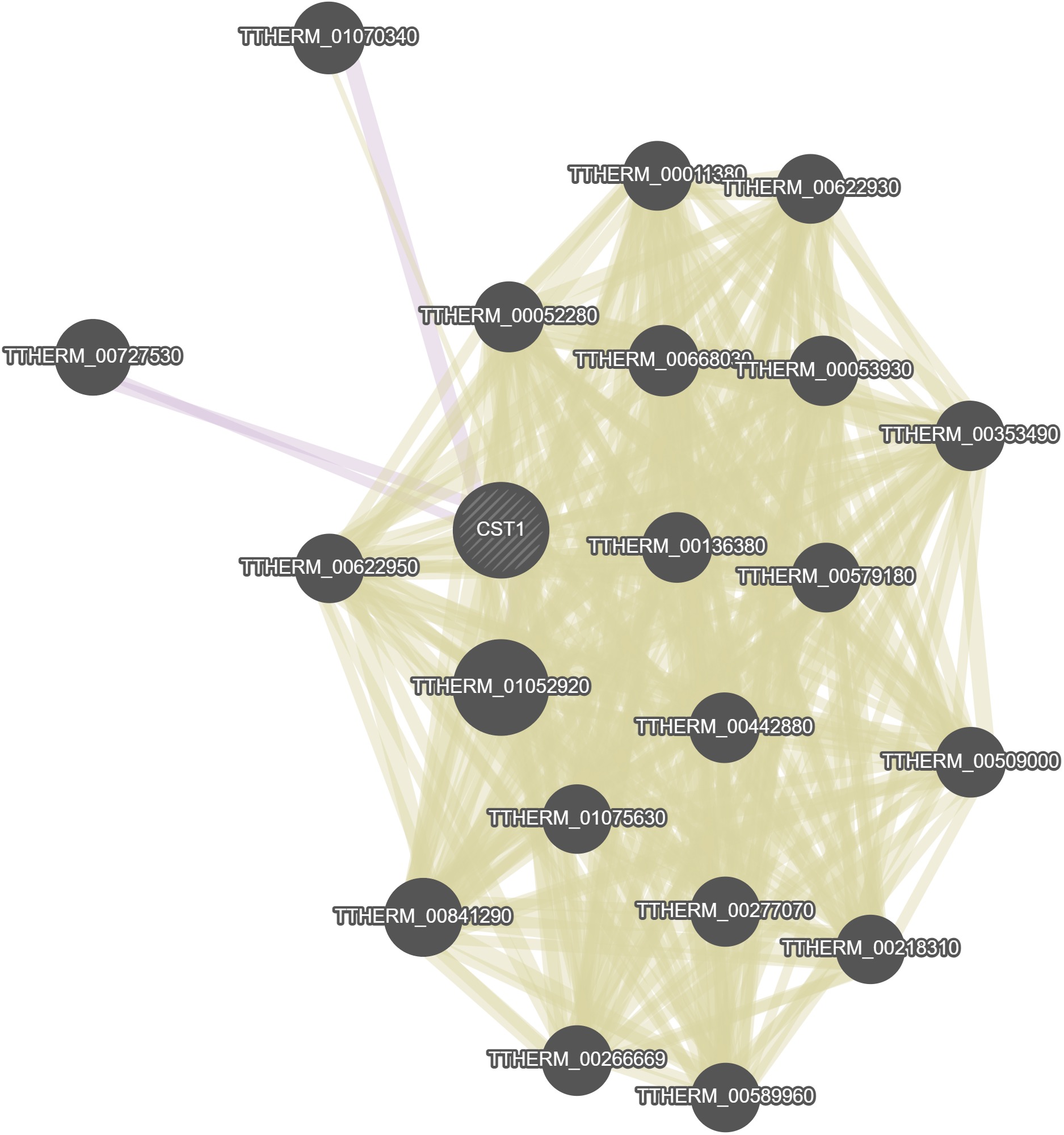
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_01014460(coding)
ATGAATCCTCTTATGAGCCCCTGTATGATTGAAAAGCAATGGCTTATTAAATCTAAAATG
GGAAATATTGAAGATGACTATGATTTTGACTAAAAGAAAGATAATTTAGGTGAAGGATCA
TATGGTGTTGTATATAAAGCTACCATGAAATCAACTGGATAAATTAGGGCTGTGAAAGTT
ATTAATAAAAGCAGAGTAAAACATTAAGAGCGCTTAAAAAATGAAATTGAAATTATGTCA
ACACTGGATCATCCTAACATTATCAGATTATATGAGACATATGAAGATCAGAAAAATATT
TACTTGGTTATAGAATACTGTGAAGGAGGAGAACTTTTTGACAGAATAGCAGAAAGAGGC
TACTATACAGAGTAAGATGCTGTTGCACTATTTAGATAAATTATGAGTGCAATCAATTAT
TGTCACTCTCGAAAAATATGTCATAGAGACTTAAAACCTGAAAATTTCTTGTTTTGCACT
AAAGATGACAACTCAATAATTAAGGTAATTGATTTTGGTCTTTCTAAAACATATGGGCAA
GTTTTAGATTCTCCTCTCAAAAAGAAGCCTTTAGAATTAACTCAAGTACAAAGCGGCGTT
ATGAAAACTAGAGCAGGAACACCATATTATATTGCTCCAGAAGTACTTGATGGTAAATAT
GATGAAAAATGTGATATCTGGTCAGCTGGTGTTACTTTGTATATTATTCTTTGCGGCTAT
CCCCCATTTTATGGAGAAACTGATAAGGAAATTCTACAAGCTGTAAAAAAAGGAGAATAT
GAATATGAAGGCGATGAATGGGATATTGTGAGCGATGAATGCAAAAACTTAATCAAAAAA
ATGCTTAGTAAGCCAGAGCTAAGATTCAGTTCAGAAATGGTTATGGCCTCCAAATGGATA
TAAAAGTATAGTCCTCCAATTAAAAATGATGCCAAAATTCCATTTTAGAAGATTAAATTA
TGGAATACTTATCCCAAGCTCAAAAAGCTAGCCCTAAGCTATATTGCTTCATAAATGAGC
ATGTTTTAGATATAACCCATAGCTAAACAATTTATTTTGATTGATAAAGACTTTGATGGC
TGCTTGACTTTATAGGAGCTCATAAATGGAACATAAGACTCTCAAAACCCAGAAAATTAA
GAAATTATTAAGCAAACATTTAAATCTTTTGTTTCATATTTACCAAATAAAATAGCATTT
ACAGATTTTGTAGCTCTCGTTATGGATTCCAGCATTTATTTGAAGGATAATATAATCCGC
CAAGCCTTTAAAATTTTTGATAAAAATAACATTGGTAAAATATCTCCAAACCATCTTAAC
GAGCTCTTTTTGGATAATGCATATTACGCAAAGAAAAAAATTGATTATTGGGTTGATATG
GTAAAATGCTGTGACTTCAAGAATGATGGAGTGATTGATTACGAAGAATTTAGCAAAATG
ATCTATGAAAATAGCGAATAAATGTAAATAAGTGCTTGA>TTHERM_01014460(gene)
AATAAACTAAAATAAAAAAACATTTAAATTAATTATATCTACTTTCTTTGAAAACACATT
TTATAGAAATGAATCCTCTTATGAGCCCCTGTATGATTGAAAAGCAATGGCTTATTAAAT
CTAAAATGGGAAATATTGAAGATGACTATGATTTTGACTAAAAGAAAGATAATTTAGGTG
AAGGATCATATGGTGTTGTATATAAAGCTACCATGAAATCAACTGGATAAATTAGGGCTG
TGAAAGTTATTAATAAAAGCAGAGTAAAACATTAAGAGCGCTTAAAAAATGAAATTGAAA
TTATGTCAACACTGGATCATCCTAACATTATCAGATTATATGAGACATATGAAGATCAGA
AAAATATTTACTTGGTTATAGAATACTGTGAAGGAGGAGAACTTTTTGACAGAATAGCAG
AAAGAGGCTACTATACAGAGTAAGATGCTGTTGCACTATTTAGATAAATTATGAGTGCAA
TCAATTATTGTCACTCTCGAAAAATATGTCATAGAGACTTAAAACCTGAAAATTTCTTGT
TTTGCACTAAAGATGACAACTCAATAATTAAGGTAATTGATTTTGGTCTTTCTAAAACAT
ATGGGCAAGTTTTAGATTCTCCTCTCAAAAAGAAGCCTTTAGAATTAACTCAAGTACAAA
GCGGCGTTATGAAAACTAGAGCAGGAACACCATATTATATTGCTCCAGAAGTACTTGATG
GTAAATATGATGAAAAATGTGATATCTGGTCAGCTGGTGTTACTTTGTATATTATTCTTT
GCGGCTATCCCCCATTTTATGGAGAAACTGATAAGGAAATTCTACAAGCTGTAAAAAAAG
GAGAATATGAATATGAAGGCGATGAATGGGATATTGTGAGCGATGAATGCAAAAACTTAA
TCAAAAAAATGCTTAGTAAGCCAGAGCTAAGATTCAGTTCAGAAATGGTTATGGCCTCCA
AATGGATATAAAAGTATAGTCCTCCAATTAAAAATGATGCCAAAATTCCATTTTAGAAGA
TTAAATTATGGAATACTTATCCCAAGCTCAAAAAGCTAGCCCTAAGCTATATTGCTTCAT
AAATGAGCATGTTTTAGATATAACCCATAGCTAAACAATTTATTTTGATTGATAAAGACT
TTGATGGCTGCTTGACTTTATAGGAGCTCATAAATGGAACATAAGACTCTCAAAACCCAG
AAAATTAAGAAATTATTAAGCAAACATTTAAATCTTTTGTTTCATATTTACCAAATAAAA
TAGCATTTACAGATTTTGTAGCTCTCGTTATGGATTCCAGCATTTATTTGAAGGATAATA
TAATCCGCCAAGCCTTTAAAATTTTTGATAAAAATAACATTGGTAAAATATCTCCAAACC
ATCTTAACGAGCTCTTTTTGGATAATGCATATTACGCAAAGAAAAAAATTGATTATTGGG
TTGATATGGTAAAATGCTGTGACTTCAAGAATGATGGAGTGATTGATTACGAAGAATTTA
GCAAAATGATCTATGAAAATAGCGAATAAATGTAAATAAGTGCTTGATATAAATTTCAAA
AATAATAATTTAAAATTTATGTTTAATAGAAAATAGTCTTTATACATACAAATAATTAAT
AATTTACATAAATAAAAACTTATTTTATTTGTTTGTTTGTTTGTTTGATTTGACTGTTTA
ATTCCTAAAAATTTAAAAACC>TTHERM_01014460(protein)
MNPLMSPCMIEKQWLIKSKMGNIEDDYDFDQKKDNLGEGSYGVVYKATMKSTGQIRAVKV
INKSRVKHQERLKNEIEIMSTLDHPNIIRLYETYEDQKNIYLVIEYCEGGELFDRIAERG
YYTEQDAVALFRQIMSAINYCHSRKICHRDLKPENFLFCTKDDNSIIKVIDFGLSKTYGQ
VLDSPLKKKPLELTQVQSGVMKTRAGTPYYIAPEVLDGKYDEKCDIWSAGVTLYIILCGY
PPFYGETDKEILQAVKKGEYEYEGDEWDIVSDECKNLIKKMLSKPELRFSSEMVMASKWI
QKYSPPIKNDAKIPFQKIKLWNTYPKLKKLALSYIASQMSMFQIQPIAKQFILIDKDFDG
CLTLQELINGTQDSQNPENQEIIKQTFKSFVSYLPNKIAFTDFVALVMDSSIYLKDNIIR
QAFKIFDKNNIGKISPNHLNELFLDNAYYAKKKIDYWVDMVKCCDFKNDGVIDYEEFSKM
IYENSEQMQISA