Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01020640Standard Name
LC4BAliases
PreTt17631 | 200.m00044 | 3756.m00240Description
LC4B dynein 18 kDa light chain; Light chain subunit of ciliary three-headed outer arm dynein (OAD); Calmodulin and Centrin Calcium-Binding FamiliesGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- axonemal dynein complex (ISS) | GO:0005858
- microtubule associated complex (ISS) | GO:0005875
- outer dynein arm (ISS) | GO:0036157
Molecular Function
- microtubule motor activity (ISS) | GO:0003777
Biological Process
- cilium movement (ISS) | GO:0003341
- microtubule-based movement (ISS) | GO:0007018
- microtubule-based process (ISS) | GO:0007017
 Domains
Domains
- ( PF13499 ) EF-hand domain pair
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
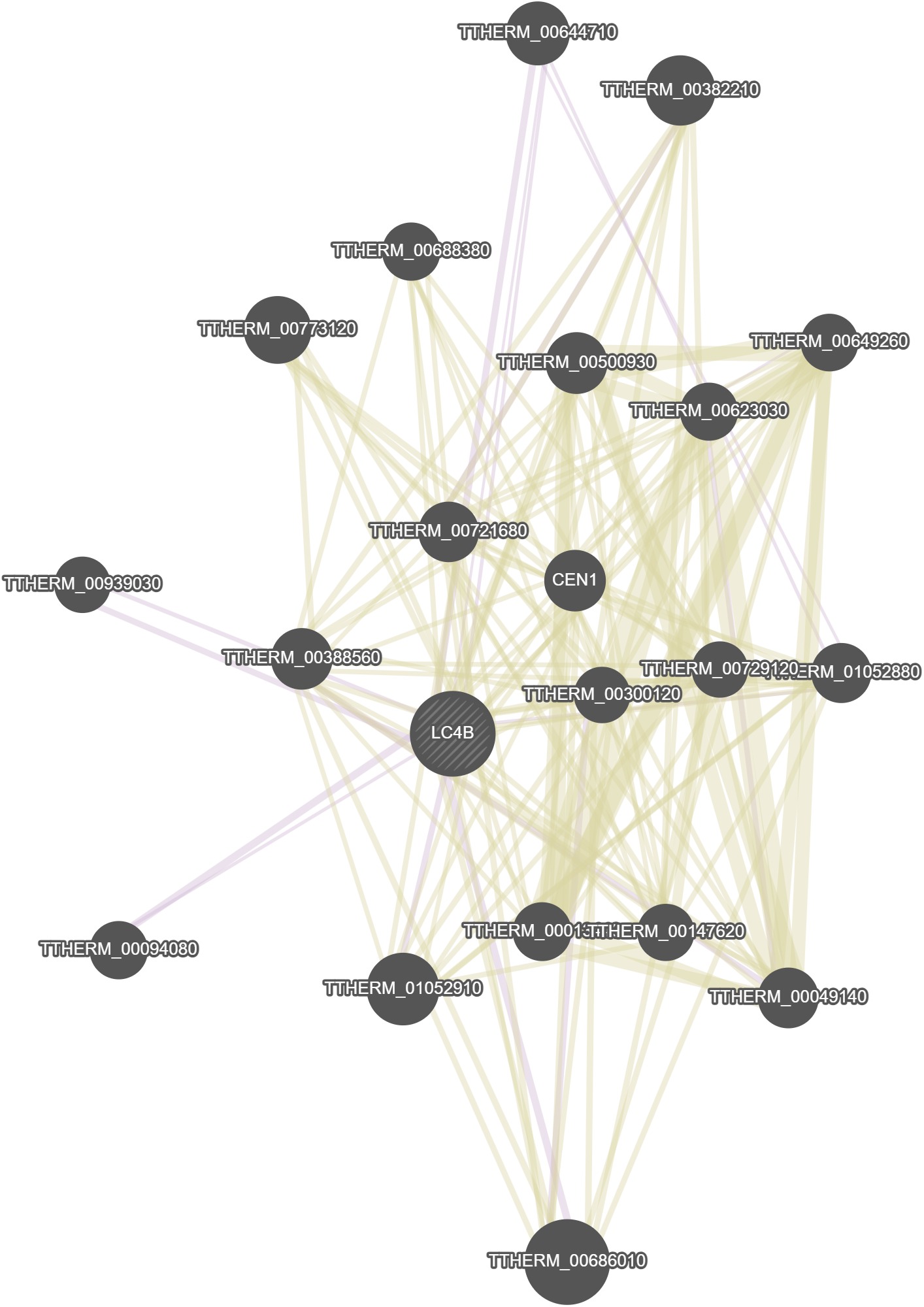
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_01020640(coding)
ATGGATAAAATAAATCCTGAAAAGATTTTAAGCGAAGAAGAAAAAAAAGCTTGCAAAGAA
TCATTTGATGCGTATGATAAATTAGGATATGGAACTTTAGAAGTAGAAGAACTTTAAAAA
GTATTAGAAGAACTAGGTGAAAAACCTACTAAGGAGGAATTATACAAAATGATAAGTGAA
GTAGATAAAACTAATAAAGGATATATAGAATTCAATGACTTTATGAGAGCAATAGCATAT
TATAAAATGCTTGAAAAAGAATCAGAAAACGATGATACATTGGAAGCTTTTGTTGCAATG
GGAGGTTCAGTTGACAAATCAGGAAACGTTGATACTGCTAAATTGATATAAATTGTAAGA
GATGAATTTAATATGACTATAGATATCGAAAGATTAATACATGAAATAGATCATGATAAG
AGTGGTAAAATATCTTTTGATGAATTTAAAGCTTTACTCTCTAAATGA>TTHERM_01020640(gene)
TAATTAGCGGTGATAAATTTTATAATTAATTTCGCAAAGTAAATATTCTTTTTATTTTCA
AGGCACAGTCAAAAAAAATCAAACAAATGGATAAAATAAATCCTGAAAAGATTTTAAGCG
AAGAAGAAAAAAAAGCTTGCAAAGAATCATTTGATGCGTATGATAAATTAGGATATGGAA
CTTTAGAAGTAGAAGAACTTTAAAAAGTATTAGAAGAACTAGGTGAAAAACCTACTAAGG
AGGAATTATACAAAATGATAAGTGAAGTAGATAAAACTAATAAAGGATATATAGAATTCA
ATGACTTTATGAGAGCAATAGCATATTATAAAATGCTTGAAAAAGAATCAGAAAACGATG
ATACATTGGAAGCTTTTGTTGCAATGGGAGGTTCAGTTGACAAATCAGGAAACGTTGATA
CTGCTAAATTGATATAAATTGTAAGAGATGAATTTAATATGACTATAGATATCGAAAGAT
TAATACATGAAATAGATCATGATAAGAGTGGTAAAATATCTTTTGATGAATTTAAAGCTT
TACTCTCTAAATGAAGGGGAATAGATAATTTCAATTCTTTGGATTTTTTTTTGTATAGTT
ATGCTGGTAGTTAACTAGAAATATTATTTTAAGAAATGTAACTAATGGGCAGATTAAAAC
ATAAAAATTTAATAAGTTATAATTAAAGTAGAATTATGAAATATATTAGTTAAAATTTAT
AGTTTAGTTATTTATATTTTTATTATCTGATTTTTAAGAGTTGATAAATTATATTTTATT
TATTTATACACTTATTTTT>TTHERM_01020640(protein)
MDKINPEKILSEEEKKACKESFDAYDKLGYGTLEVEELQKVLEELGEKPTKEELYKMISE
VDKTNKGYIEFNDFMRAIAYYKMLEKESENDDTLEAFVAMGGSVDKSGNVDTAKLIQIVR
DEFNMTIDIERLIHEIDHDKSGKISFDEFKALLSK