Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01079200Standard Name
HTA4 (Histone Two A 4)Aliases
PreTt17435 | 221.m00055 | 3832.m00156 | HTAYDescription
HTA4 HTAY histone H2A; Core histone H2A variant; Histone H2AGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- cellulosome (IEA) | GO:0043263
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- obsolete ectoplasm (IEA) | GO:0043265
- prospore membrane spindle pole body attachment site (IEA) | GO:0070057
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-keto-3-deoxy-L-rhamnonate aldolase activity (IEA) | GO:0106099
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- CAP-Gly domain binding (IEA) | GO:0071794
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- inositol monophosphate 1-phosphatase activity (IEA) | GO:0008934
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lanosterol synthase activity (IEA) | GO:0000250
- lipopolysaccharide-1,6-galactosyltransferase activity (IEA) | GO:0008921
- mannitol-1-phosphate 5-dehydrogenase activity (IEA) | GO:0008926
- mannose-1-phosphate guanylyltransferase (GDP) activity (IEA) | GO:0008928
- mitochondrial single subunit type RNA polymerase activity (IEA) | GO:0001065
- obsolete bacterial-type RNA polymerase activity (IEA) | GO:0001061
- obsolete murein DD-endopeptidase activity (IEA) | GO:0008931
- obsolete plastid PEP-B RNA polymerase activity (IEA) | GO:0001063
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- proteinase activated receptor binding (IEA) | GO:0031871
- siderophore-iron transmembrane transporter activity (IEA) | GO:0015343
- type 1 proteinase activated receptor binding (IEA) | GO:0031872
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to dopamine (IEA) | GO:1903351
- cellular response to homocysteine (IEA) | GO:1905375
- coenzyme A biosynthetic process (IEA) | GO:0015937
- coenzyme A metabolic process (IEA) | GO:0015936
- donor selection (IEA) | GO:0007535
- extrinsic apoptotic signaling pathway via death domain receptors (IEA) | GO:0008625
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- germ-line processes downstream of sex determination signal (IEA) | GO:0007547
- glycolytic process from glycerol (IEA) | GO:0061613
- glycolytic process through fructose-6-phosphate (IEA) | GO:0061615
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- inactivation of recombination (HML) (IEA) | GO:0007537
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- lateral sprouting from an epithelium (IEA) | GO:0060601
- learning (IEA) | GO:0007612
- learning or memory (IEA) | GO:0007611
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- lung saccule development (IEA) | GO:0060430
- maltoheptaose catabolic process (IEA) | GO:2001123
- mesenchymal cell differentiation involved in kidney development (IEA) | GO:0072161
- metanephric mesenchymal cell differentiation (IEA) | GO:0072162
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of anterior head development (IEA) | GO:2000743
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete aging (IEA) | GO:0007568
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete maintenance of dosage compensation (IEA) | GO:0007551
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete positive regulation of transcription from RNA polymerase II promoter involved in stress response to cadmium ion (IEA) | GO:1902901
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of nucleic acid-templated transcription (IEA) | GO:1903506
- obsolete reproduction (IEA) | GO:0000003
- obsolete response to long exposure to lithium ion (IEA) | GO:0043460
- ommochrome biosynthetic process (IEA) | GO:0006727
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of proteasomal protein catabolic process (IEA) | GO:1901800
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of stomatal opening (IEA) | GO:1902458
- primary lung bud formation (IEA) | GO:0060431
- pronephric glomerulus morphogenesis (IEA) | GO:0035775
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine nucleotide salvage (IEA) | GO:0032262
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of bile acid metabolic process (IEA) | GO:1904251
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of ecdysteroid secretion (IEA) | GO:0007555
- regulation of endothelial cell chemotaxis (IEA) | GO:2001026
- regulation of gastric motility (IEA) | GO:1905333
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of plant epidermal cell differentiation (IEA) | GO:1903888
- regulation of RNA binding transcription factor activity (IEA) | GO:1905255
- response to cell cycle checkpoint signaling (IEA) | GO:0072396
- sesquiterpenoid biosynthetic process (IEA) | GO:0016106
- somatic processes downstream of sex determination signal (IEA) | GO:0007546
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- striated muscle myosin thick filament assembly (IEA) | GO:0071688
- synaptic vesicle priming (IEA) | GO:0016082
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- transcription by RNA polymerase V (IEA) | GO:0001060
- tRNA import into mitochondrion (IEA) | GO:0016031
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
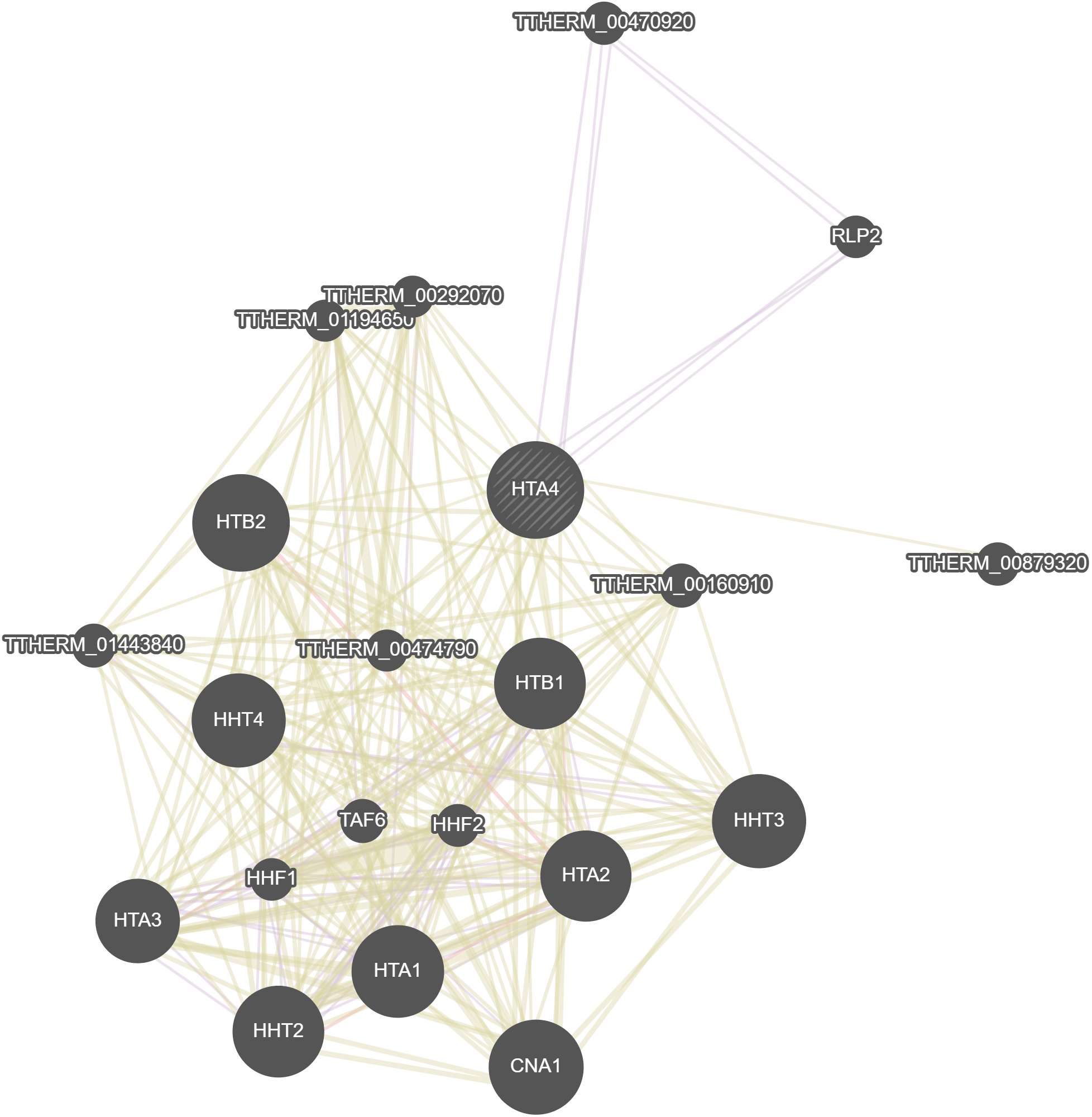
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
- ( SD01769 ) Macronucleus: Flag C-terminal tagged HTAY gene fragment coding for AA123 to 388 into MTT1 locus
- ( SD01786 ) Macronucleus: Neo3 KO of HTAY(may be incomplete) and HTAY with two HA tags on the N-terminus inserted into MTT1 locus with neo2 cassete in 5' flanking region
- ( SD01997 ) Macronucleus: Neo3 KO of HTAY (may be incomplete) and HTAY with two HA tags on the N-terminus inserted into MTT1 locus with neo2 cassete in 5' flanking region
- ( SD02200 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02201 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02202 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02203 ) Macronucleus: Neo3 into HTAY, Chimeric YN-HTAX into MTT1 locus
- ( SD02368 ) Macronucleus: 2 HA tagged HTAY into MTT1 with neo2 in 5' flank
- ( SD02381 ) Micronucleus: Neo3 replaces bases coding for AA258-388 Macronucleus: Neo3 replaces bases coding for AA258-388
- ( SD02676 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene Macronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02677 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene Macronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02678 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02679 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02680 ) Micronucleus: Neo3 ko of last 2/3 (AA123-388) of HTAY gene
- ( SD02700 ) Macronucleus: 3HA n-terminal tagged HTAY gene fragment coding for AA123 to 388into MTT1 locus
- ( SD02701 ) Macronucleus: Flag tagged 2/3 c-terminus of HTAY into MTT1 locus
- ( SD02808 ) Macronucleus: Partial ko of HTAY coding with neo3
- ( SD02809 ) Macronucleus: Partial ko of HTAY coding with neo3
- ( SD02881 ) Macronucleus: Partial replacement of MTT1 coding by N-terminus of HTAY, aa 1-122, with neo2 in 5’ flank of MTT1
- ( SD02882 ) Macronucleus: Region of HTAY gene encoding AA 123 to 257 with two HA tags at n-terminus was inserted into the MTT1 gene.
- ( SD02883 ) Macronucleus: Region of HTAY gene encoding AA 258 to 388 with two HA tags at n-terminus was inserted into the MTT1 gene.
- ( SD02884 ) Macronucleus: Region of HTAY gene encoding AA 1 to 258 with two HA tags at n-terminus was inserted into the MTT1 gene.
- ( SD02885 ) Macronucleus: Region of HTAY gene encoding AA 1 to 122 and 258 to 388 with two HA tags at n-terminus was inserted into the MTT1 gene.
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:22426537: Song X, Bowen J, Miao W, Liu Y, Gorovsky MA (2012) The nonhistone, N-terminal tail of an essential, chimeric H2A variant regulates mitotic H3-S10 dephosphorylation. Genes & development 26(6):615-29
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
 Sequences
Sequences
>TTHERM_01079200(coding)
ATGTCAACTGACTAAGAAAAGATAATAAAATAAGTATTAGATTCAATAGCAGAGCAATAG
GGAGATAGTAGAGATGATGAAGTTTATGTGATAGAAATAATGTCATTACGCTTTGAGAAA
TTTGATAATCGAATTAAGTAGAAGATTGAAAGATTTACATCTCTTTAAATGCTAACAATC
AACGATTGCCTTATCTCTGATTTAACTAACTTCCCTCATGTTCCTTCTTTGATTAGATTA
GACTTGGTCTTTAATAAAATTACTGGAGACTAGCTTTAATATTTGAGAGGATCAAGACAT
TTACAAACTTTAATGCTTGGTGCAAATTAAATTGAAGAGATCGAAGACTTGAAGAGACTC
GGTTAGATGAGAGAATTAATTCAACTAGATTTATTGAATAATCCAGTTGTTAATACAAAT
AATTATAGAAATTTAGTTTTTAATTTGTTTCCTTCTTTGGTAATATTAGACACTTTAGAT
AAAAATGGAATTGATTAAGAAAAAGCAGCTCTTGATATTTCTGCTTCAAGAGTGCCTGAT
AACTTATTTGATAAATCTAAGCCTGTATAAAATGTTTCCAAATAAGTAGTTAAAAATGCT
AAAGCTACTTAAAATAATCTTTTCAGTTCTACACAAAAAGTAGCTTAAGTTAAATAAATT
CCCAAAATTGTACCTGCAAAAGTATCTAAGCCTTCTCAAGCATCAGTTTAAGACAACAAA
TCTAATGTAGTTTAAGCAAAAGTTACTGCTGCTGTAAGTGTTGGTAGAAAAGCACCTGCA
TCAAGAAATGGTGGAGTACCATCAAAAGCTGGAAAAGGCAAAATGAATGCTTTTTCAAAG
TAGTCTACATCTCAAAAAAGTGGTTTAGTATTTCCTTGTGGAAGACTTAGAAGATACTTG
AAACAGGCTTGCAAACAATTTAGAGTTTCTTCAAGCTGTAACATTTACTTAGCTGGTGTT
CTTGAATACTTAGCAGCAGAAGTCTTAGAAACTGCAGGAAATATAGCAAAAAACAATAGG
CTAAGTAGAATTAACCCAGTTCATATAAGAGAAGCTTTTAGAAACGATGCTGAGTTAAAC
TAATTTGTAAATGGTACTATTATAGCTGAAGGAGGAAGCACAAGTACTAATTTTGTCCTT
CCAATTATTAATAAATCCAAAAAATGA>TTHERM_01079200(gene)
ATAGTATAAATAAAAAACTTATATATACAAATAAAGATATTAATAATAACTTTGACAAAC
ATTTTTACAAAAAATAAAAAATAAGTTAAATAAACACAACAATAATCTTTAAAAATGTCA
ACTGACTAAGAAAAGATAATAAAATAAGTATTAGATTCAATAGCAGAGCAATAGGGAGAT
AGTAGAGATGATGAAGTTTATGTGATAGAAATAATGTCATTACGCTTTGAGAAATTTGAT
AATCGAATTAAGTAGAAGATTGAAAGATTTACATCTCTTTAAATGCTAACAATCAACGAT
TGCCTTATCTCTGATTTAACTAACTTCCCTCATGTTCCTTCTTTGATTAGATTAGACTTG
GTCTTTAATAAAATTACTGGAGACTAGCTTTAATATTTGAGAGGATCAAGACATTTACAA
ACTTTAATGCTTGGTGCAAATTAAATTGAAGAGATCGAAGACTTGAAGAGACTCGGTTAG
ATGAGAGAATTAATTCAACTAGATTTATTGAATAATCCAGTTGTTAATACAAATAATTAT
AGAAATTTAGTTTTTAATTTGTTTCCTTCTTTGGTAATATTAGACACTTTAGATAAAAAT
GGAATTGATTAAGAAAAAGCAGCTCTTGATATTTCTGCTTCAAGAGTGCCTGATAACTTA
TTTGATAAATCTAAGCCTGTATAAAATGTTTCCAAATAAGTAGTTAAAAATGCTAAAGCT
ACTTAAAATAATCTTTTCAGTTCTACACAAAAAGTAGCTTAAGTTAAATAAATTCCCAAA
ATTGTACCTGCAAAAGTATCTAAGCCTTCTCAAGCATCAGTTTAAGACAACAAATCTAAT
GTAGTTTAAGCAAAAGTTACTGCTGCTGTAAGTGTTGGTAGAAAAGCACCTGCATCAAGA
AATGGTGGAGTACCATCAAAAGCTGGAAAAGGCAAAATGAATGCTTTTTCAAAGTAGTCT
ACATCTCAAAAAAGTGGTTTAGTATTTCCTTGTGGAAGACTTAGAAGATACTTGAAACAG
GCTTGCAAACAATTTAGAGTTTCTTCAAGCTGTAACATTTACTTAGCTGGTGTTCTTGAA
TACTTAGCAGCAGAAGTCTTAGAAACTGCAGGAAATATAGCAAAAAACAATAGGCTAAGT
AGAATTAACCCAGTTCATATAAGAGAAGCTTTTAGAAACGATGCTGAGTTAAACTAATTT
GTAAATGGTACTATTATAGCTGAAGGAGGAAGCACAAGTACTAATTTTGTCCTTCCAATT
ATTAATAAATCCAAAAAATGATTAAAAAAAAAATTCAAGTTAAATAAAATATAAAAATAA
AAATAATCACTAAATAATTTATCAATATAAAATAATTTAAAAATAAATATAGTTTATAAT
ATTAATAAATAAATAAATAAATAAAATAATCATTAACATTATTTTCATTTTATTGTCATT
TTATTTCCAAATTTAAGTCCTAAAGT>TTHERM_01079200(protein)
MSTDQEKIIKQVLDSIAEQQGDSRDDEVYVIEIMSLRFEKFDNRIKQKIERFTSLQMLTI
NDCLISDLTNFPHVPSLIRLDLVFNKITGDQLQYLRGSRHLQTLMLGANQIEEIEDLKRL
GQMRELIQLDLLNNPVVNTNNYRNLVFNLFPSLVILDTLDKNGIDQEKAALDISASRVPD
NLFDKSKPVQNVSKQVVKNAKATQNNLFSSTQKVAQVKQIPKIVPAKVSKPSQASVQDNK
SNVVQAKVTAAVSVGRKAPASRNGGVPSKAGKGKMNAFSKQSTSQKSGLVFPCGRLRRYL
KQACKQFRVSSSCNIYLAGVLEYLAAEVLETAGNIAKNNRLSRINPVHIREAFRNDAELN
QFVNGTIIAEGGSTSTNFVLPIINKSKK