Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01142730Standard Name
RPL9 (Ribosomal protein of the large subunit)Aliases
PreTt04328 | 252.m00039 | 3824.m03216 | RPL6-E.coliDescription
RPL9 60S ribosomal protein L9; Ribosomal protein L6 containing protein; Homolog of yeast RPL9(/RPL6) and E. coli RPL6; Large ribosomal subunit protein uL6-likeGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 11beta-hydroxyprogesterone binding (IEA) | GO:1903879
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- CCR10 chemokine receptor binding (IEA) | GO:0031735
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- prenyltransferase activity (IEA) | GO:0004659
- protein histidine kinase activity (IEA) | GO:0004673
Biological Process
- asymmetric protein localization (IEA) | GO:0008105
- basement membrane organization (IEA) | GO:0071711
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- coenzyme A metabolic process (IEA) | GO:0015936
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- microtubule depolymerization (IEA) | GO:0007019
- mitochondrial membrane organization (IEA) | GO:0007006
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of glycoprotein metabolic process (IEA) | GO:1903019
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of stomatal opening (IEA) | GO:1902457
- negative regulation of telomere single strand break repair (IEA) | GO:1903824
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete bent DNA binding (IEA) | GO:0006326
- obsolete larval behavior (sensu Insecta) (IEA) | GO:0007627
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- pigment accumulation in response to UV light (IEA) | GO:0043478
- pigment accumulation in tissues in response to UV light (IEA) | GO:0043479
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of B cell mediated immunity (IEA) | GO:0002714
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of peroxisome proliferator activated receptor signaling pathway (IEA) | GO:0035360
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of wound healing, spreading of epidermal cells (IEA) | GO:1903691
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of glycoprotein metabolic process (IEA) | GO:1903018
 Domains
Domains
- ( PF00347 ) Ribosomal protein L6
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
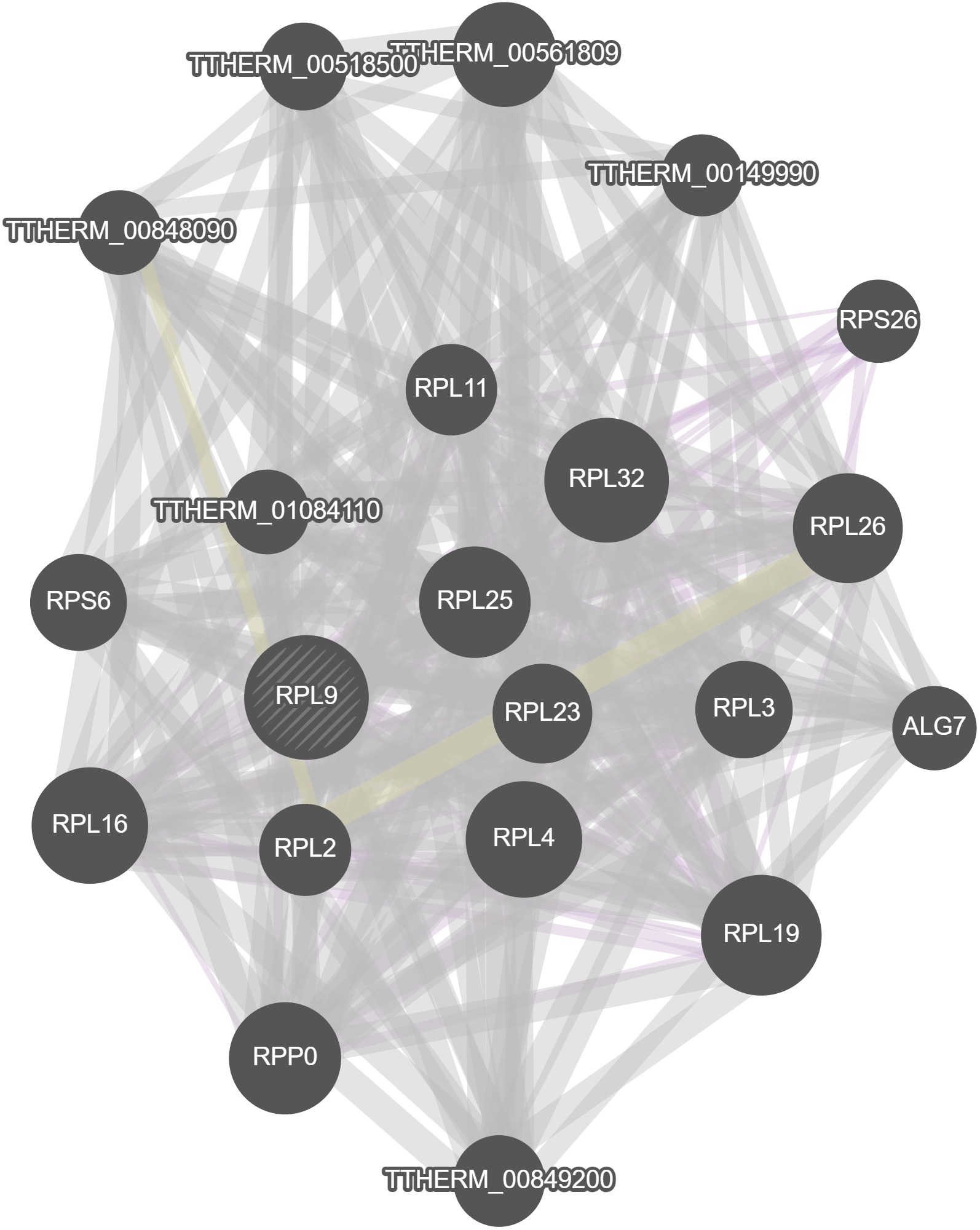
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
- Ref:22052974: Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N (2011) Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science (New York, N.Y.) 334(6058):941-8
 Sequences
Sequences
>TTHERM_01142730(coding)
ATGAGACACTTACTTACCGAAGTCAACGTACCCATCCCTGATAAGGTCACCATCACTGCT
AAATAAAGAGTAGTTGAAGTCAAGGGACCCTTAGGTACCATCAAGAGAGCTTTCAGATAC
GCCTCTGTTGATATTCAAAAGCCTACCGCCGACAACGTTAAGTTATAAATCTGGTAAGCT
TCCAGAAAGGAAAGAGCCGTCCTCCAATCTATTGCTTCTCAAATCAAGAATATGATTAGA
GGTGTTACTGAAGGTTACAAATTCAAGATGAAGCTTGCCTTCGCTCACTTCCCCATTTAA
GAAGCTGTTGCTAAGGATGGTTCCTCCATTGAAATTAAGCATTTCTTGGGTGAAAAGAGA
ATTAGAAGAATCCAAGCTCTTCCTGGTGTCAAGATCTCCAGAAAGGACGAAGAAAAGAAT
ACCCTCACCCTCTAAGGTATTGACTTGAACAACGTCTCCCAAACTTGCGCTCTCATCCAC
CAATCTTGTTTGGTCAAAGAAAAGGATATCAGACAATTCTTGGACGGTATTTACGTTTCT
GATAAGAGACTCGCCATGAACGAGTGA>TTHERM_01142730(gene)
TTTCATTCATTTTAGAATGAAAAATGTTTTCATTTTATTTTGATTGAAAATCGGATGTAT
AAAATAAAGACAAGAAGAAATTAAAAATAAAGGCAAAAAGTAAGAGTCAAATTAATAAAA
CAAAAAAGTTAGAAATAATAAACAAAAAAGAAAATTAATAGGATATTAAAAAAAATATTA
AAGTAAAAATGAGACACTTACTTACCGAAGTCAACGTACCCATCCCTGATAAGGTCACCA
TCACTGCTAAATAAAGAGTAGTTGAAGTCAAGGGACCCTTAGGTACCATCAAGAGAGCTT
TCAGATACGCCTCTGTTGATATTCAAAAGCCTACCGCCGACAACGTTAAGTTATAAATCT
GGTAAGCTTCCAGAAAGGAAAGAGCCGTCCTCCAATCTATTGCTTCTCAAATCAAGAATA
TGATTAGAGGTGTTACTGAAGGTTACAAATTCAAGATGAAGCTTGCCTTCGCTCACTTCC
CCATTTAAGAAGCTGTTGCTAAGGATGGTTCCTCCATTGAAATTAAGCATTTCTTGGGTG
AAAAGAGAATTAGAAGAATCCAAGCTCTTCCTGGTGTCAAGATCTCCAGAAAGGACGAAG
AAAAGAATACCCTCACCCTCTAAGGTATTGACTTGAACAACGTCTCCCAAACTTGCGCTC
TCATCCACCAATCTTGTTTGGTCAAAGAAAAGGATATCAGACAATTCTTGGACGGTATTT
ACGTTTCTGATAAGAGACTCGCCATGAACGAGTGATTTAATAAATCTACTATAAAATATA
TACTAAAATTTGATGTTATTATTGCAAACATTTTAATTTGTATAAATATAAATTTTTGTA
AATCAAAT>TTHERM_01142730(protein)
MRHLLTEVNVPIPDKVTITAKQRVVEVKGPLGTIKRAFRYASVDIQKPTADNVKLQIWQA
SRKERAVLQSIASQIKNMIRGVTEGYKFKMKLAFAHFPIQEAVAKDGSSIEIKHFLGEKR
IRRIQALPGVKISRKDEEKNTLTLQGIDLNNVSQTCALIHQSCLVKEKDIRQFLDGIYVS
DKRLAMNE