Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01161020Standard Name
NIT2 (NITrilase superfamily)Aliases
PreTt26356 | 259.m00040 | 3823.m01241Description
NIT2 carbon-nitrogen family hydrolase; hydrolase, carbon-nitrogen family protein; NITrilase superfamily; Carbon-nitrogen hydrolaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- MalFGK2 complex (IEA) | GO:1990176
- nuclear inner membrane (IEA) | GO:0005637
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- aromatase activity (IEA) | GO:0008402
- cyclase activator activity (IEA) | GO:0010853
- Edg-7 lysophosphatidic acid receptor binding (IEA) | GO:0031760
- follicle-stimulating hormone receptor binding (IEA) | GO:0031762
- galanin receptor binding (IEA) | GO:0031763
- lysine:proton symporter activity (IEA) | GO:0015493
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- peptide-glutamate-alpha-N-acetyltransferase activity (IEA) | GO:1990190
- phosphatidylinositol-3-phosphate binding (IEA) | GO:0032266
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- putrescine:ornithine antiporter activity (IEA) | GO:0015496
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sterol 14-demethylase activity (IEA) | GO:0008398
- type 2 galanin receptor binding (IEA) | GO:0031765
Biological Process
- asymmetric cell division (IEA) | GO:0008356
- autoinducer AI-2 transmembrane transport (IEA) | GO:1905887
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to osmotic stress (IEA) | GO:0071470
- external genitalia morphogenesis (IEA) | GO:0035261
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- interleukin-12 production (IEA) | GO:0032615
- interleukin-17 production (IEA) | GO:0032620
- interleukin-19 production (IEA) | GO:0032622
- interleukin-2 production (IEA) | GO:0032623
- internal genitalia morphogenesis (IEA) | GO:0035260
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of hemopoiesis (IEA) | GO:1903707
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of pseudohyphal growth (IEA) | GO:2000221
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of tumor necrosis factor-mediated signaling pathway (IEA) | GO:0010804
- nuclear division (IEA) | GO:0000280
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete positive regulation of mitotic G1 cell cycle arrest in response to nitrogen starvation (IEA) | GO:1903694
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of siRNA processing (IEA) | GO:1903705
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- response to rest involved in regulation of muscle adaptation (IEA) | GO:0014893
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- sodium-independent thromboxane transport (IEA) | GO:0071721
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- vesicle cargo loading (IEA) | GO:0035459
 Domains
Domains
- ( PF00795 ) Carbon-nitrogen hydrolase
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
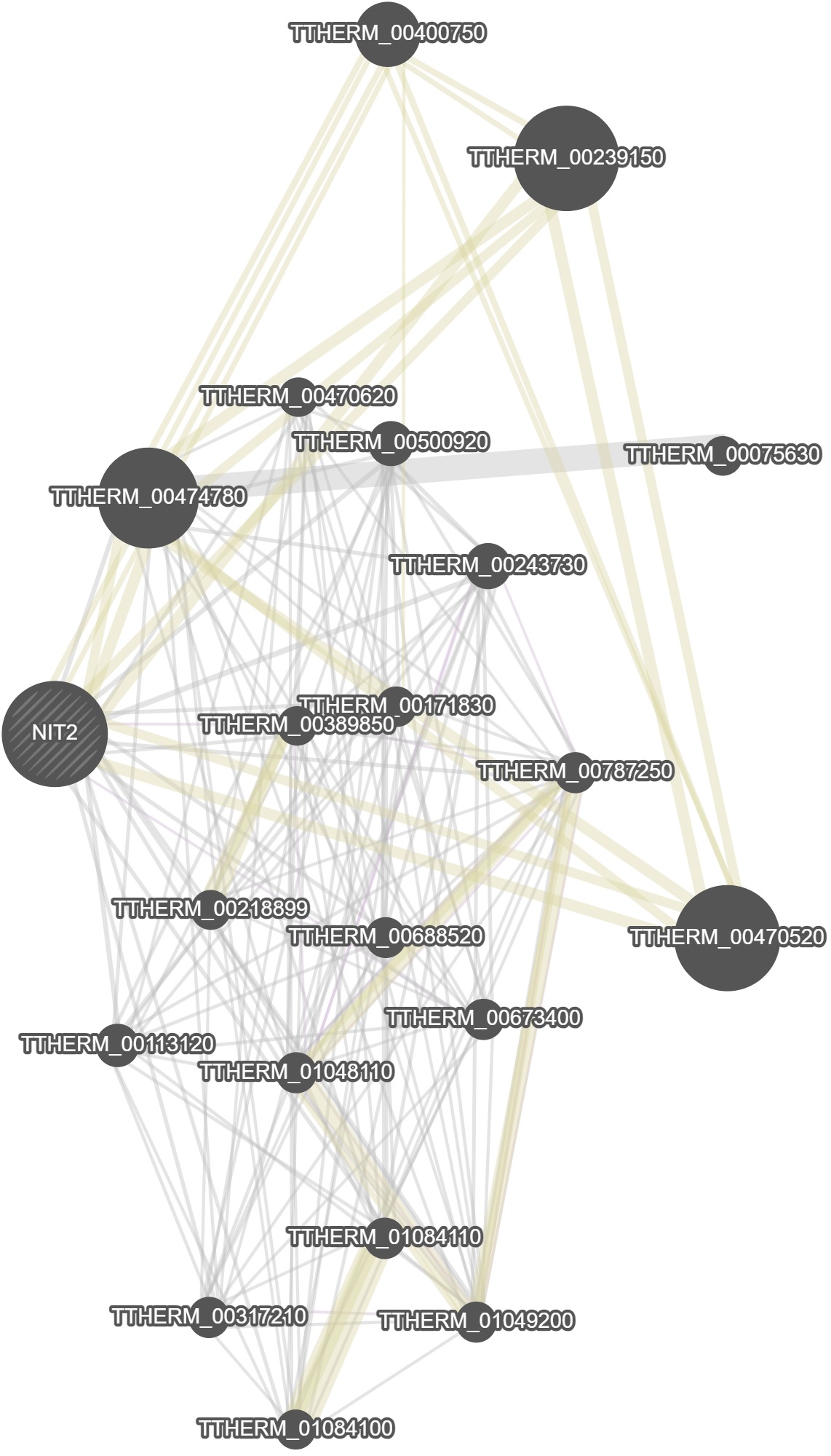
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No Data fetched for General Information
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_01161020(coding)
ATGAATCAAATTAGAAAATGCTTAGTAGGTGTTGTTTAGATGTGTTCTACTCACAATAAA
AAATAAAATATGGAATTTATTCTTTAAAATTTAAAATAAGCTCATGAAAAATAAGCGAAA
ATATGCTTTTTTCCTGAAGCCTTTGCAATGATTTCTAGAAGTTTTGCAGAAACATTTGAA
AACGCAGAATATATTGATGGTGAAATGATAAATTGCTTAAGAGATCATGCTAAAAAGTAT
AATTTGTGGCTATCGCTTGGAGGCTTTTAGGAAAGATTAAAAGAAAATGATAAAAAGATG
GGAAATACTCATATCATTATTGATAATTTAGGAAACATAGTATAAACTTATAAGAAGCTT
CATCTTTTTGATATAAGCATCGACACAAAAAACACAATATCTGAAAGTAGTGGTTATGTA
TTTGGCGACTAAGTTCCAAATGTTGTTGACTCTCCTGCAGGCAGGTTAGGATTATCTATT
TGCTATGACTTGAGATTCCCTGAGCTTTTTAGACTTCTCGCAGTTCAACAGAAAGCTGAA
ATATTATTAGTTCCAAGTGCTTTTTTTAAAAAGACTGGCTAAGCTCACTGGCATACTCTA
CTAAAAGCTAGAGCCATAGAAAATTAATGCTTCGTTATTGCAGCTGCATAAGCAGGATAG
CATAATGATAAAAGAGAAAGCTATGGACACTCTTTAGTTATAGATCCTTGGGGTGAAGTA
TTATTGGATATGGGACCAGAAAAGCTTGGTATTGATTTTGTAGAAATAGATTTGGAAAAA
ATTGATACAGTCAGAAAAGGTTTACCAGCTATAAATCATATTAGAAACGATATTTACAAG
CTATCTAAAATTTGA>TTHERM_01161020(gene)
TAAATCTTGAAAAAATAACATCAAAATTAATAAAATAAAGATAGAAGTTTTCAATTTTAC
AATATAGTTTGAATAAATATCATGAATCAAATTAGAAAATGCTTAGTAGGTGTTGTTTAG
ATGTGTTCTACTCACAATAAAAAATAAAATATGGAATTTATTCTTTAAAATTTAAAATAA
GCTCATGAAAAATAAGCGAAAATATGCTTTTTTCCTGAAGCCTTTGCAATGATTTCTAGA
AGTTTTGCAGAAACATTTGAAAACGCAGAATATATTGATGGTGAAATGATAAATTGCTTA
AGAGATCATGCTAAAAAGTATAATTTGTGGCTATCGCTTGGAGGCTTTTAGGAAAGATTA
AAAGAAAATGATAAAAAGATGGGAAATACTCATATCATTATTGATAATTTAGGAAACATA
GTATAAACTTATAAGAAGCTTCATCTTTTTGATATAAGCATCGACACAAAAAACACAATA
TCTGAAAGTAGTGGTTATGTATTTGGCGACTAAGTTCCAAATGTTGTTGACTCTCCTGCA
GGCAGGTTAGGATTATCTATTTGCTATGACTTGAGATTCCCTGAGCTTTTTAGACTTCTC
GCAGTTCAACAGAAAGCTGAAATATTATTAGTTCCAAGTGCTTTTTTTAAAAAGACTGGC
TAAGCTCACTGGCATACTCTACTAAAAGCTAGAGCCATAGAAAATTAATGCTTCGTTATT
GCAGCTGCATAAGCAGGATAGCATAATGATAAAAGAGAAAGCTATGGACACTCTTTAGTT
ATAGATCCTTGGGGTGAAGTATTATTGGATATGGGACCAGAAAAGCTTGGTATTGATTTT
GTAGAAATAGATTTGGAAAAAATTGATACAGTCAGAAAAGGTTTACCAGCTATAAATCAT
ATTAGAAACGATATTTACAAGCTATCTAAAATTTGATAAAAAATATTTATTTGGATTTTT
ATAAGCAAGCAACAAATATAAATAATTATATTGTAATTTATT>TTHERM_01161020(protein)
MNQIRKCLVGVVQMCSTHNKKQNMEFILQNLKQAHEKQAKICFFPEAFAMISRSFAETFE
NAEYIDGEMINCLRDHAKKYNLWLSLGGFQERLKENDKKMGNTHIIIDNLGNIVQTYKKL
HLFDISIDTKNTISESSGYVFGDQVPNVVDSPAGRLGLSICYDLRFPELFRLLAVQQKAE
ILLVPSAFFKKTGQAHWHTLLKARAIENQCFVIAAAQAGQHNDKRESYGHSLVIDPWGEV
LLDMGPEKLGIDFVEIDLEKIDTVRKGLPAINHIRNDIYKLSKI