Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01243450Standard Name
OGL1 (OGG1-Like)Aliases
PreTt01756 | 287.m00027Description
OGL1 HhH-GPD family base excision DNA repair protein; HhH-GPD superfamily base excision DNA repair protein; Oxidative DNA damage repair enzymeGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- alpha6-beta1 integrin-CD151 complex (IEA) | GO:0071059
- classical-complement-pathway C3/C5 convertase complex (IEA) | GO:0005601
- complement component C1 complex (IEA) | GO:0005602
- EGFR-Grb2-Sos complex (IEA) | GO:0070620
- gamma-catenin-TCF7L2 complex (IEA) | GO:0071665
- insulin-responsive compartment (IEA) | GO:0032593
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- karyomere (IEA) | GO:0061468
- MalFGK2 complex (IEA) | GO:1990176
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- obsolete plasma membrane part of cell junction (IEA) | GO:0061466
- obsolete septin patch (IEA) | GO:0032167
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- amino acid transmembrane transporter activity (IEA) | GO:0015171
- aromatase activity (IEA) | GO:0008402
- cAMP-dependent protein kinase regulator activity (IEA) | GO:0008603
- cyclase activator activity (IEA) | GO:0010853
- DNA demethylase activity (IEA) | GO:0035514
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- lysine:proton symporter activity (IEA) | GO:0015493
- methane monooxygenase [NAD(P)H] activity (IEA) | GO:0015049
- microtubule severing ATPase activity (IEA) | GO:0008568
- mitochondrial RNA polymerase binding promoter specificity activity (IEA) | GO:0000998
- MutSbeta complex binding (IEA) | GO:0032408
- obsolete 4,5-dihydroxypyrene dioxygenase activity (IEA) | GO:0034922
- obsolete cation:cation antiporter activity (IEA) | GO:0015491
- obsolete myosin ATPase activity (IEA) | GO:0008570
- obsolete nucleoplasmin ATPase activity (IEA) | GO:0008572
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete proprotein convertase 2 activator activity (IEA) | GO:0016484
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete type II protein secretor activity (IEA) | GO:0015447
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- oligoxyloglucan reducing-end-specific cellobiohydrolase activity (IEA) | GO:0033945
- peptide-glutamate-alpha-N-acetyltransferase activity (IEA) | GO:1990190
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphoglycerate mutase activity (IEA) | GO:0004619
- purinergic nucleotide receptor activity (IEA) | GO:0001614
- putrescine:ornithine antiporter activity (IEA) | GO:0015496
- pyrimidine nucleotide-sugar transmembrane transporter activity (IEA) | GO:0015165
- RNA polymerase II transcription regulatory region sequence-specific DNA binding (IEA) | GO:0000977
- RNA polymerase III type 3 promoter sequence-specific DNA binding (IEA) | GO:0001006
- silver ion transmembrane transporter activity (IEA) | GO:0015080
- single-subunit type RNA polymerase binding (IEA) | GO:0001050
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- sterol 14-demethylase activity (IEA) | GO:0008398
- titin binding (IEA) | GO:0031432
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- uridine phosphorylase activity (IEA) | GO:0004850
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- anterior midgut development (IEA) | GO:0007496
- apoptotic program (IEA) | GO:0008632
- asymmetric cell division (IEA) | GO:0008356
- autoinducer AI-2 transmembrane transport (IEA) | GO:1905887
- basement membrane organization (IEA) | GO:0071711
- basolateral protein localization (IEA) | GO:0061467
- cell surface receptor signaling pathway involved in heart development (IEA) | GO:0061311
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to desiccation (IEA) | GO:0071465
- cellular response to homocysteine (IEA) | GO:1905375
- cellular response to osmotic stress (IEA) | GO:0071470
- cellulose microfibril organization (IEA) | GO:0010215
- clypeo-labral disc morphogenesis (IEA) | GO:0007453
- coenzyme A biosynthetic process (IEA) | GO:0015937
- cytotoxic T cell pyroptotic cell death (IEA) | GO:1902483
- endoderm development (IEA) | GO:0007492
- endodermal cell fate determination (IEA) | GO:0007493
- establishment or maintenance of polarity of embryonic epithelium (IEA) | GO:0016332
- ethylene biosynthetic process (IEA) | GO:0009693
- external genitalia morphogenesis (IEA) | GO:0035261
- fractalkine production (IEA) | GO:0032603
- gamma-tubulin complex assembly (IEA) | GO:1902481
- glycolytic process from glycerol (IEA) | GO:0061613
- granulocyte macrophage colony-stimulating factor production (IEA) | GO:0032604
- hair cycle process (IEA) | GO:0022405
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- hemocyte development (IEA) | GO:0007516
- homogentisate metabolic process (IEA) | GO:1901999
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- induction of blood coagulation in another organism (IEA) | GO:0035807
- interleukin-35 biosynthetic process (IEA) | GO:0070749
- interleukin-5 production (IEA) | GO:0032634
- internal genitalia morphogenesis (IEA) | GO:0035260
- laminaritriose transport (IEA) | GO:2001097
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- lung saccule development (IEA) | GO:0060430
- macrophage derived foam cell differentiation (IEA) | GO:0010742
- maintenance of imaginal histoblast diploidy (IEA) | GO:0007489
- maltotetraose transport (IEA) | GO:2001099
- mesodermal cell fate determination (IEA) | GO:0007500
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of cardiac muscle tissue regeneration (IEA) | GO:1905179
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine (C-C motif) ligand 20 production (IEA) | GO:1903885
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of endopeptidase activity (IEA) | GO:0010951
- negative regulation of epithelial cell-cell adhesion involved in epithelium migration (IEA) | GO:1903682
- negative regulation of hemopoiesis (IEA) | GO:1903707
- negative regulation of intestinal phytosterol absorption (IEA) | GO:0010949
- negative regulation of macrophage derived foam cell differentiation (IEA) | GO:0010745
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of postsynaptic membrane organization (IEA) | GO:1901627
- negative regulation of sprouting angiogenesis (IEA) | GO:1903671
- negative regulation of telomeric D-loop disassembly (IEA) | GO:1905839
- negative regulation of the force of heart contraction by chemical signal (IEA) | GO:0003108
- negative regulation of tumor necrosis factor-mediated signaling pathway (IEA) | GO:0010804
- neutral lipid metabolic process (IEA) | GO:0006638
- nitrate transmembrane transport (IEA) | GO:0015706
- obsolete aging (IEA) | GO:0007568
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete MAPK cascade involved in ascospore formation (IEA) | GO:1903695
- obsolete mRNA endonucleolytic cleavage involved in unfolded protein response (IEA) | GO:0070055
- obsolete positive regulation of heparan sulfate proteoglycan biosynthesis by positive regulation of epimerase activity (IEA) | GO:0010910
- obsolete positive regulation of mitotic G1 cell cycle arrest in response to nitrogen starvation (IEA) | GO:1903694
- obsolete regulation of 1-deoxy-D-xylulose-5-phosphate synthase activity (IEA) | GO:1902395
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete sodium/potassium transport (IEA) | GO:0006815
- ommochrome biosynthetic process (IEA) | GO:0006727
- pericardial cell differentiation (IEA) | GO:0007513
- phagocytosis (IEA) | GO:0006909
- phagocytosis, engulfment (IEA) | GO:0006911
- photoreceptor cell morphogenesis (IEA) | GO:0008594
- photosynthesis, light harvesting in photosystem I (IEA) | GO:0009768
- phyllotactic patterning (IEA) | GO:0060771
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of establishment of RNA localization to telomere (IEA) | GO:1904912
- positive regulation of macrophage derived foam cell differentiation (IEA) | GO:0010744
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of response to water deprivation (IEA) | GO:1902584
- positive regulation of siRNA processing (IEA) | GO:1903705
- positive regulation of thyroid gland epithelial cell proliferation (IEA) | GO:1904443
- positive regulation of transporter activity (IEA) | GO:0032411
- primary branching, open tracheal system (IEA) | GO:0007428
- programmed cell death (IEA) | GO:0012501
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein K27-linked deubiquitination (IEA) | GO:1990167
- protein localization to mitotic spindle (IEA) | GO:1902480
- protein localization to old growing cell tip (IEA) | GO:1903858
- prothoracic disc morphogenesis (IEA) | GO:0007470
- pyrimidine deoxyribonucleotide biosynthetic process (IEA) | GO:0009221
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of ascospore wall beta-glucan biosynthetic process (IEA) | GO:0060622
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of establishment of Sertoli cell barrier (IEA) | GO:1904444
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of glucose metabolic process (IEA) | GO:0010906
- regulation of L-arginine import into cell (IEA) | GO:1902826
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of protein localization to cell division site (IEA) | GO:1901900
- regulation of satellite cell activation involved in skeletal muscle regeneration (IEA) | GO:0014717
- regulation of sprouting angiogenesis (IEA) | GO:1903670
- response to bacterial lipopeptide (IEA) | GO:0070339
- response to cycloalkane (IEA) | GO:0014071
- response to interleukin-11 (IEA) | GO:0071105
- siderophore metabolic process (IEA) | GO:0009237
- sodium-independent prostaglandin transport (IEA) | GO:0071720
- sodium-independent thromboxane transport (IEA) | GO:0071721
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- synaptic vesicle priming (IEA) | GO:0016082
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- tumor necrosis factor production (IEA) | GO:0032640
- vesicle cargo loading (IEA) | GO:0035459
- visceral mesoderm-endoderm interaction involved in midgut development (IEA) | GO:0007495
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
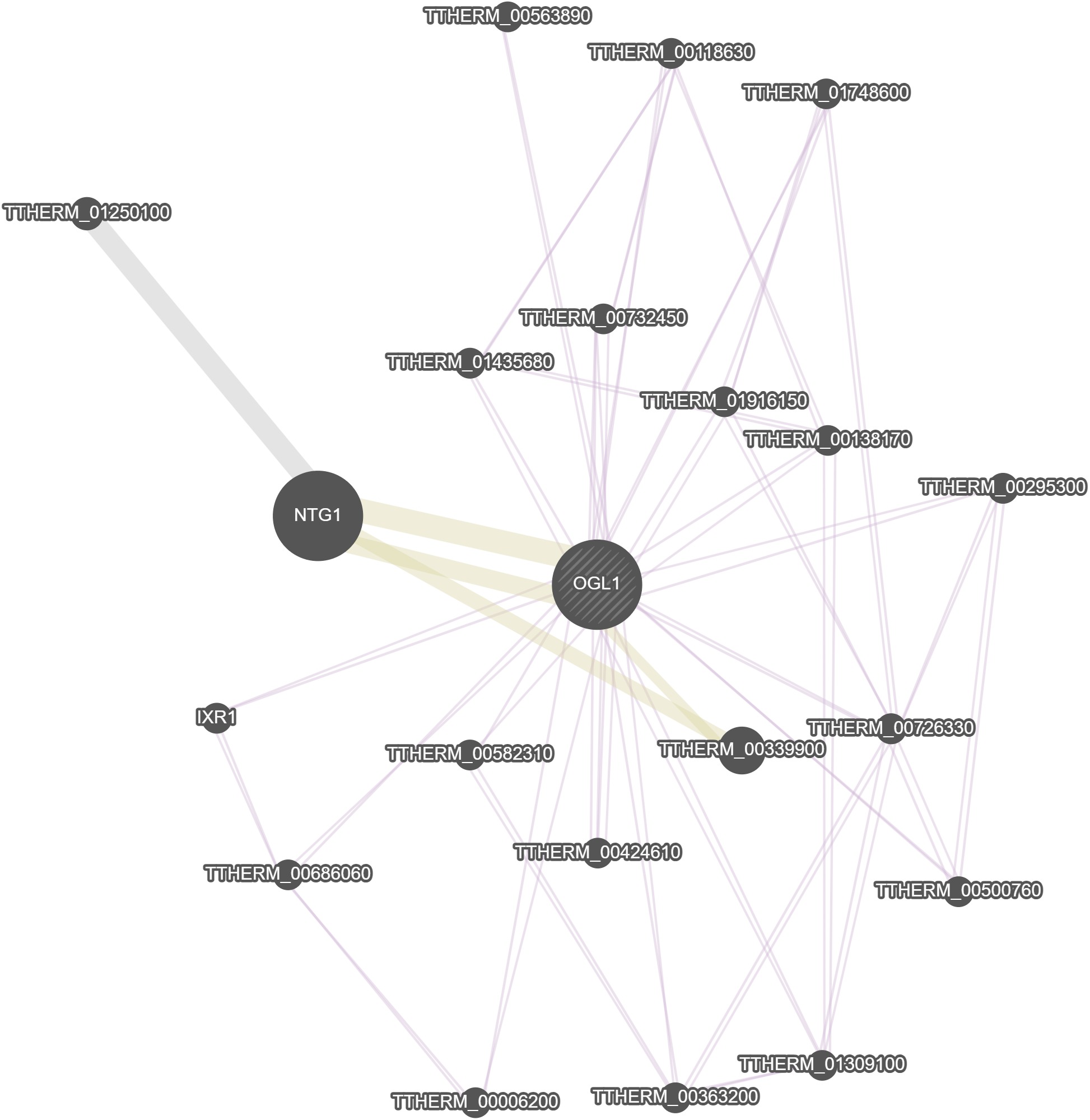
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2294 OGL1 Tetrahymena thermophila’s OGL1 protein is homologous to proteins from the HhH-GPD and OGG-N superfamilies, which are associated with DNA repair. It contains two domains; the first domain (E-value= 3.0 -17), in the HhH-GPD family, is a Helix-hairpin-helix and Gly/Pro rich loop, found on proteins that remove base lesions. The second domain (E-value=6.9-13), is common to oxoguanine gylcosylases, also known as OGG1, which removes 8-oxoG lesions. The closest homologs are in Scheffersomyces stipitis, a negative Crabtree yeast, and in Taphrina deformans, a fungi/plant pathogen, with 46% identical residues. OGL1 may be an essential protein in preventing mutations in DNA that lead to a short lifespan, early aging, and cancer. Paragraph by: Lillian Horin, Maite Cortes Garcia, Francis Ryu, Forrest Fulgenzi, Keck Science Center, Pitzer College
 Associated Literature
Associated Literature
No Data fetched for Associated Literature
 Sequences
Sequences
>TTHERM_01243450(coding)
ATGAAGTCTCTAATAAACCCCTTAACTAAAAAGCCAATAACATAGATAGAATTCAATTTA
AAGAATACCTTGGTCAACGGCCAAGCATTTAATTGGATAAACAAAGATGTAACTTATAAC
AAAGAGTTTAGAGGGATTCTAGGTAAAAATGTAGTAGATTTTAAAGTTAATGAAAATAAT
GAAGTAATGTACAAAGAAATACCAGAAAATTCAAAAATTGAGGAGATTATTAATGATTAC
TTGCAATTAAATGTAGATTTAGTCAAGCTTACTGTTGAGTGGTCAGAAAAAGATAACTAT
TATGCTAAAGTCAAAGAATAGTTAAAAGGAGTTCGTATCTTGAGATAGTTTCCATTTGAG
TGCATGATTAGCTTTATATGTTCCTAAAACAATAATATTCCAAGAATTACTAAAATTTTA
AAAAGTTTGAGATAGAATTTTGGAGAGGAAATTTATAGAGAAAAATTAGAAGATGGTAGC
GAAGAAATTTATTATAGTTTTCCATCTGTAGCATCATTGAGGAAAGCAACAGAACAAAAT
CTCAGAGATTTGGGATTGGGTTATAGAGCTAATTATATTGTCTCAGCAGTTTAGCTACTT
GAGGAGAAAGGAGGAGAAGAATATTTGCACAAATTGCGCTAGCTGAAAGATTCACATGAG
AAAAGAGAAGCCCTGCTTGAATTCAAAGGAATAGGAAATAAAGTAGCAGATTGTATATCT
CTCTTTAGCTTAGATGCAAAAGATTTAATCCCTGTAGATACTCATGTTTTTCAAATATAT
AATAGAGTTTACAAAAAAAATAAAAACAAAGATAAAAAAATGACAAAAGATCAGTATTCA
GAAATAGAAAATTTCTTTAAAGATCTATATGGAGATTATGCAGGATGGGCCCATCAAGCT
ATTTTTGCGGCTGAACTCGTCTCTTTTTAGTCAGAAATTAACTCAAACGAAAACTCTTCA
GTAAAAAAAACAAAATCATCAAAATTACAAGAGGAAAAAGAATAAGAAAAAGAATAAACT
ACAGAGGAGTAAAAAATTATTTCTAAAAGTAAAATTAGCGAATAAACATCTTCAGAGAAA
AATAAAAGAAATTAAAACAAAAAAATAAAGTGA>TTHERM_01243450(gene)
TTTGAATTTAATTTTGAGTGTTTATAAATCTGTAAAGTAGAAGTGATTAGAATTTTGTTG
ATTTAGCAATTTATATCTATTAGGCTCTCTATTTATATATTTTGATGCCAAAAATAATAA
TTATCTTCTTTTTATTAAATAATTTAATTATCATTAAATATTATGAAGTCTCTAATAAAC
CCCTTAACTAAAAAGCCAATAACATAGATAGAATTCAATTTAAAGAATACCTTGGTCAAC
GGCCAAGCATTTAATTGGATAAACAAAGATGTAACTTATAACAAAGAGTTTAGAGGGATT
CTAGGTAAAAATGTAGTAGATTTTAAAGTTAATGAAAATAATGAAGTAATGTACAAAGAA
ATACCAGAAAATTCAAAAATTGAGGAGATTATTAATGATTACTTGCAATTAAATGTAGAT
TTAGTCAAGCTTACTGTTGAGTGGTCAGAAAAAGATAACTATTATGCTAAAGTCAAAGAA
TAGTTAAAAGGAGTTCGTATCTTGAGATAGTTTCCATTTGAGTGCATGATTAGCTTTATA
TGTTCCTAAAACAATAATATTCCAAGAATTACTAAAATTTTAAAAAGTTTGAGATAGAAT
TTTGGAGAGGAAATTTATAGAGAAAAATTAGAAGATGGTAGCGAAGAAATTTATTATAGT
TTTCCATCTGTAGCATCATTGAGGAAAGCAACAGAACAAAATCTCAGAGATTTGGGATTG
GGTTATAGAGCTAATTATATTGTCTCAGCAGTTTAGCTACTTGAGGAGAAAGGAGGAGAA
GAATATTTGCACAAATTGCGCTAGCTGAAAGATTCACATGAGAAAAGAGAAGCCCTGCTT
GAATTCAAAGGAATAGGAAATAAAGTAGCAGATTGTATATCTCTCTTTAGCTTAGATGCA
AAAGATTTAATCCCTGTAGATACTCATGTTTTTCAAATATATAATAGAGTTTACAAAAAA
AATAAAAACAAAGATAAAAAAATGACAAAAGATCAGTATTCAGAAATAGAAAATTTCTTT
AAAGATCTATATGGAGATTATGCAGGATGGGCCCATCAAGCTATTTTTGCGGCTGAACTC
GTCTCTTTTTAGTCAGAAATTAACTCAAACGAAAACTCTTCAGTAAAAAAAACAAAATCA
TCAAAATTACAAGAGGAAAAAGAATAAGAAAAAGAATAAACTACAGAGGAGTAAAAAATT
ATTTCTAAAAGTAAAATTAGCGAATAAACATCTTCAGAGAAAAATAAAAGAAATTAAAAC
AAAAAAATAAAGTGAAAAAGTTTTAATTTATGTTATATTTAATTGATACTTAAATGGATT
ACAAAAAGTAAACATACAAGAAACTTTAAATTTTTTTATAAGTAGAAAATAGAAAATAAA
TAAAAAAAAAATAATTTAATAAAATATTCCTGAATTAATCTTCAATAATATTAGGTTAAA
ATATATAAATAATAAATATTTACTAATTAAAAAAAAATTTTTATTTGTAAAAATTCTATT
TATTTTTATAGATAAATTTATTTATTTTAATTTAAATAAGTTAAAATACATTTATGTTCA
TTATAAG>TTHERM_01243450(protein)
MKSLINPLTKKPITQIEFNLKNTLVNGQAFNWINKDVTYNKEFRGILGKNVVDFKVNENN
EVMYKEIPENSKIEEIINDYLQLNVDLVKLTVEWSEKDNYYAKVKEQLKGVRILRQFPFE
CMISFICSQNNNIPRITKILKSLRQNFGEEIYREKLEDGSEEIYYSFPSVASLRKATEQN
LRDLGLGYRANYIVSAVQLLEEKGGEEYLHKLRQLKDSHEKREALLEFKGIGNKVADCIS
LFSLDAKDLIPVDTHVFQIYNRVYKKNKNKDKKMTKDQYSEIENFFKDLYGDYAGWAHQA
IFAAELVSFQSEINSNENSSVKKTKSSKLQEEKEQEKEQTTEEQKIISKSKISEQTSSEK
NKRNQNKKIK