Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00125280Standard Name
PDD1 (Programmed DNA Degradation 1)Aliases
PreTt09290 | 9.m00311 | 3699.m00021Description
PDD1 chromodomain protein; Chromodomain protein involved in DNA elimination from the developing macronuclear genome; absent from vegetative and starved cells, but strongly enriched in developing macronuclei; co-localizes with micronuclear-limited DNA sequences; Heterochromatin-associated chromo domain-containing proteinGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- heterochromatin (IDA) | GO:0000792
- macronucleus (IDA) | GO:0031039
- micronucleus (IDA) | GO:0031040
Molecular Function
- methylated histone binding (IDA) | GO:0035064
Biological Process
- programmed DNA elimination (IMP) | GO:0031049
- scnRNA processing (IMP) | GO:0031051
 Domains
Domains
- ( PF00385 ) Chromo (CHRromatin Organisation MOdifier) domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
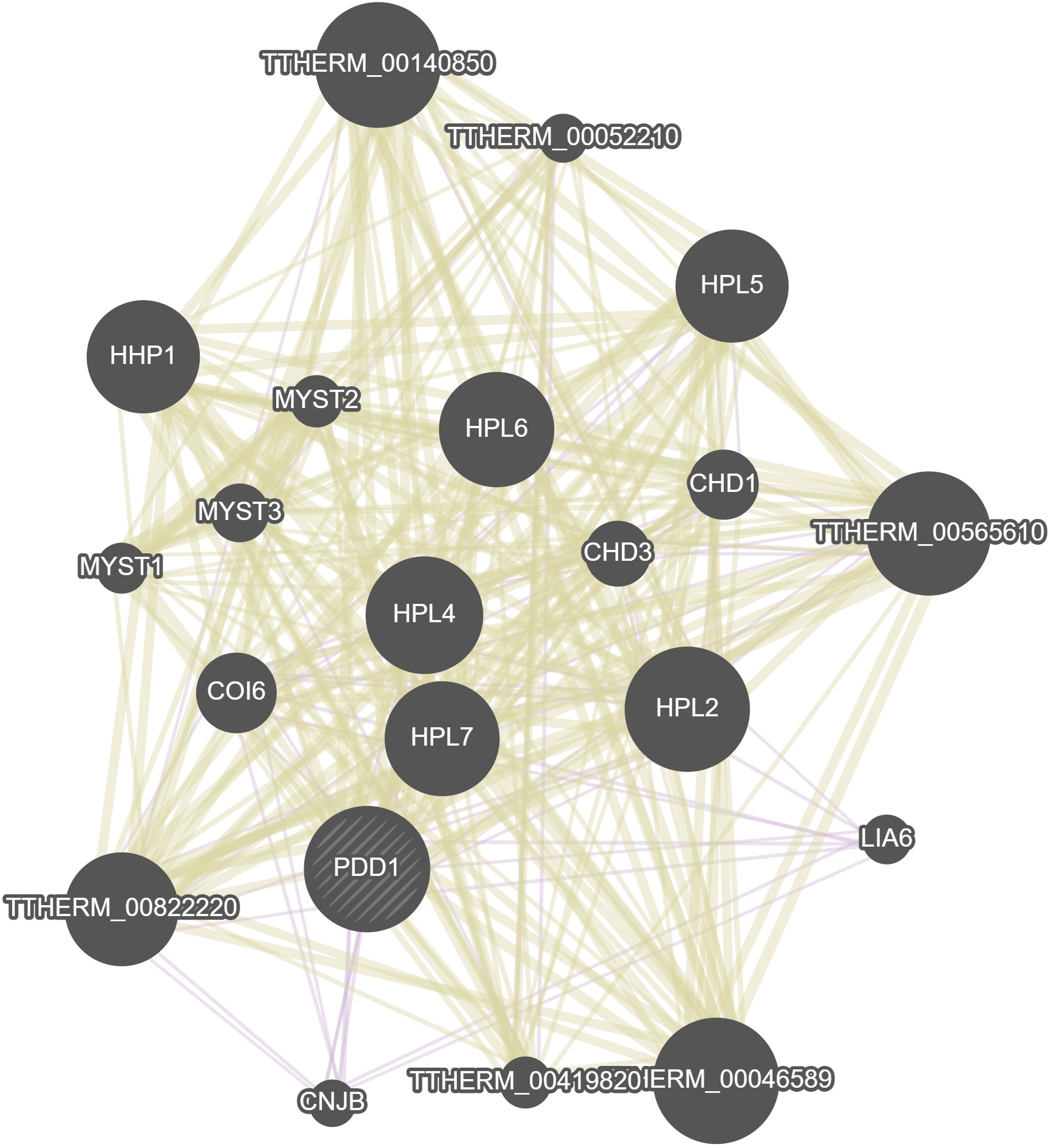
No. Gene Name(s) Paragraph Text 2209 PDD1, HHT2, DCL1, PDD3, TWI1 A proposed model for the mechanism of programmed DNA elimination in Tetrahymena is based on the timing of expression, cellular distribution, mutant phenotypes, and predicted functions of the protein and RNA components involved. In this model, both strands of the micronuclear genome (or perhaps only the portions containing internal eliminated sequences) are transcribed early in conjugation to produce large non-genic, double-stranded RNAs. This transcription is likely performed by RNA Polymerase II, based on the localization of its subunit Rpb3p to the micronucleus during this time. These transcripts pass to the cytoplasm where they are processed into short (~28 nucleotide) scan RNAs (scnRNA) by the dicer-like protein Dcl1p, similar to the production of the small inhibitory RNAs (siRNA) central to the RNA interference (RNAi) pathway of other eukaryotes. The scnRNAs complex with Twi1p, a member of the PPD (PAZ and Piwi Domain) protein family, whose members are commonly involved in RNAi and related processes. The scnRNA/Twi1p complexes enter the old macronucleus, where scnRNAs homologous to DNA sequences found there are degraded. The remaining scnRNAs, comprised of micronuclear-restricted sequences, are transferred to the developing macronucleus. There, histone H3 proteins (Hht1p, Hht2p) that are bound to sections of the genome sharing identity to the scnRNAs are methylated on lysine-9. This modification, which is often associated with the formation of heterochromatin, is recognized by one or more of the chromodomains belonging to Pdd1p and Pdd3p. Regions of DNA associated with these modified histones are eliminated from the developing macronuclear genome. 2208 PDD1 The PDD1 gene encodes Pdd1p, an abundant protein whose expression is limited to the sexual phase of the Tetrahymena life cycle. Somatic knockout cells lacking Pdd1p during the early stages of conjugation and macronuclear development exhibit defects in a variety of developmental processes, including programmed DNA elimination, macronuclear genome endoduplication, and nuclear resorption. While Pdd1p is not required for vegetative growth, exconjugants derived from matings of somatic knockout cells are inviable. Originally identified during a screen for proteins upregulated during macronuclear development (which also led to the cloning of PDD2 and PDD3), the gene encoding p65 (Pdd1p) was cloned and shown to encode a novel protein composed of three chromodomains. Methylated histone binding activity has been demonstrated in vitro for one chromodomain of Pdd1p, specifically to methylated lysine-9 residues of histone H3. This histone modification is required for programmed DNA elimination, and like Pdd1p, these modified histones colocalize with chromatin containing the DNA sequences destined for elimination. Distribution of Pdd1p in the cell over time follows a remarkable pattern that is suggestive of its major role in programmed DNA elimination: Pdd1p is initially restricted to the old macronucleus, then relocalizes to the developing macronucleus when it is formed. Studies have long suggested an epigenetic contribution from the parental macronucleus that specifies the elimination of specific DNA sequences. The timing of its localization and its ability to bind chromatin suggests Pdd1p is directly involved in communicating this information to the new macronucleus. 2316 JUB1, PDD1 Jub1p promotes heterochromatin body formation and dephosphorylation of the Heterochromatin Protein 1-like protein Pdd1p. Because Pdd1p dephosphorylation promotes the electrostatic interaction between Pdd1p and RNA in vitro and heterochromatin body formation in vivo, it has been proposed that heterochromatin body is assembled by the Pdd1p-RNA interaction.
 Associated Literature
Associated Literature
- Ref:29021213: Noto T, Mochizuki K (2017) Whats, hows and whys of programmed DNA elimination in Tetrahymena. Open biology 7(10):
- Ref:28692189: Wiley EA, Horrell S, Yoshino A, C Schornak C, Bagnani C, L Chalker D (2017) Diversification of HP1-like Chromo Domain Proteins in Tetrahymena thermophila. The Journal of eukaryotic microbiology ( ):
- Ref:28273462: Suhren JH, Noto T, Kataoka K, Gao S, Liu Y, Mochizuki K (2017) Negative Regulators of an RNAi-Heterochromatin Positive Feedback Loop Safeguard Somatic Genome Integrity in Tetrahymena. Cell reports 18(10):2494-2507
- Ref:27466409: Kataoka K, Noto T, Mochizuki K (2016) Phosphorylation of an HP1-like protein is a prerequisite for heterochromatin body formation in Tetrahymena DNA elimination. Proceedings of the National Academy of Sciences of the United States of America 113(32):9027-32
- Ref:26688337: Kataoka K, Mochizuki K (2015) Phosphorylation of an HP1-like Protein Regulates Heterochromatin Body Assembly for DNA Elimination. Developmental cell 35(6):775-88
- Ref:24297443: Schwope RM, Chalker DL (2014) Mutations in Pdd1 reveal distinct requirements for its chromodomain and chromoshadow domain in directing histone methylation and heterochromatin elimination. Eukaryotic cell 13(2):190-201
- Ref:20357003: Cheng CY, Vogt A, Mochizuki K, Yao MC (2010) A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Molecular biology of the cell 21(10):1753-62
- Ref:18706458: Chalker DL (2008) Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochimica et biophysica acta 1783(11):2130-6
- Ref:17519286: Yao MC, Yao CH, Halasz LM, Fuller P, Rexer CH, Wang SH, Jain R, Coyne RS, Chalker DL (2007) Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. Journal of cell science 120(Pt 12):1978-89
- Ref:17586719: Rexer CH, Chalker DL (2007) Lia1p, a novel protein required during nuclear differentiation for genome-wide DNA rearrangements in Tetrahymena thermophila. Eukaryotic cell 6(8):1320-9
- Ref:17575054: Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD (2007) RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes & development 21(12):1530-45
- Ref:16896218: Fillingham JS, Garg J, Tsao N, Vythilingum N, Nishikawa T, Pearlman RE (2006) Molecular genetic analysis of an SNF2/brahma-related gene in Tetrahymena thermophila suggests roles in growth and nuclear development. Eukaryotic cell 5(8):1347-59
- Ref:14755052: Liu Y, Mochizuki K, Gorovsky MA (2004) Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America 101(6):1679-84
- Ref:12297043: Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA (2002) Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110(6):689-99
- Ref:12351775: Jenuwein T (2002) Molecular biology. An RNA-guided pathway for the epigenome. Science (New York, N.Y.) 297(5590):2215-8
- Ref:12297044: Taverna SD, Coyne RS, Allis CD (2002) Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110(6):701-11
- Ref:12455963: Duharcourt S, Yao MC (2002) Role of histone deacetylation in developmentally programmed DNA rearrangements in Tetrahymena thermophila. Eukaryotic cell 1(2):293-303
- Ref:10805754: Nikiforov MA, Gorovsky MA, Allis CD (2000) A novel chromodomain protein, pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Molecular and cellular biology 20(11):4128-34
- Ref:10619033: Coyne RS, Nikiforov MA, Smothers JF, Allis CD, Yao MC (1999) Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Molecular cell 4(5):865-72
- Ref:10473642: Maercker C, Kortwig H, Nikiforov MA, Allis CD, Lipps HJ (1999) A nuclear protein involved in apoptotic-like DNA degradation in Stylonychia: implications for similar mechanisms in differentiating and starved cells. Molecular biology of the cell 10(9):3003-14
- Ref:10198282: Janetopoulos C, Cole E, Smothers JF, Allis CD, Aufderheide KJ (1999) The conjusome: a novel structure in Tetrahymena found only during sexual reorganization. Journal of cell science 112 ( Pt 7)( ):1003-11
- Ref:9109258: Smothers JF, Madireddi MT, Warner FD, Allis CD (1997) Programmed DNA degradation and nucleolar biogenesis occur in distinct organelles during macronuclear development in Tetrahymena. The Journal of eukaryotic microbiology 44(2):79-88
- Ref:9409671: Smothers JF, Mizzen CA, Tubbert MM, Cook RG, Allis CD (1997) Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development (Cambridge, England) 124(22):4537-45
- Ref:9196044: Callebaut I, Courvalin JC, Worman HJ, Mornon JP (1997) Hydrophobic cluster analysis reveals a third chromodomain in the Tetrahymena Pdd1p protein of the chromo superfamily. Biochemical and biophysical research communications 235(1):103-7
- Ref:8858150: Madireddi MT, Coyne RS, Smothers JF, Mickey KM, Yao MC, Allis CD (1996) Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell 87(1):75-84
- Ref:7958410: Madireddi MT, Davis MC, Allis CD (1994) Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Developmental biology 165(2):418-31
 Sequences
Sequences
>TTHERM_00125280(coding)
ATGTTTACTGTGAAATCACTCTAATAATTTGTAAATAATTATTTTAAATCCAACCAAATA
TTTTGTTAATTTTATAAAAATAATAAATTTAAAAATTCTCCTAATTAGTAAGCTAAAAAA
GCAATAAATATTTCAATAAAGAAAATGTCTCAGAAAAAGAGTTTAAAATAAAAGAGAAAG
TAAGACTATAGCTCTGATACTGAAGAGGAAGAAGAGGATCAATATGAAGTTGAAAAGATC
TTAGACTCTAGATTTAATCCTAAGACCAAGCAAAAAGAATATCTTGTCAAATGGGAAAAC
TGGCCTATTGAAGACTCTACCTGGGAACCTTACGAGCATCTTTCGAATGTCAAAGAAATA
GTTTAGGCCTTTGAAAAGAAGCAAAAAGCTAACGTTATGCCTCAACCTACTGGACCTATC
ACTAGAGTAAATGCCTAGAAAGACCCTTAAAAAAAGAACAGACTATCTTTGAATTCTTCT
ATCAGCAAATCTTTACCTCAAGAAGAAGAAATTTAGACATCTAAAGAAGACAGTAAAAAA
TAAGCAGTCAAAAAATTCCAACCAGCTTCAAGAAGAAAATCTATTAGTTCTTAATCTGAT
TCAGACCTTTAAGCTGAAGAAGTTCCACAGTAGCCTGAAAGTAAAAAAGATAAAAATGAT
GGTGCCTTTGAAGAACCCAACAATGCTGATGCAGACGAAACTGAGCTTGTATTTGAAGAA
ATTGTTGACAAAAGAATTCTTGATGGATAAACAGAATATTTAATACGCTTCTAAAATGTC
TCTTAGCCATAATGGGTAGATGTTGGACAATTGATAGCTATTAAAGATGATGTTATAGCT
TATGAAGATAAAATTGCTGCTTAGAGTTAGCTCAATAAGACTAAGAACTTGAAAGAAAAT
GCTAAACATTCATCTTAACAATAGCAAAGCGAAAATAATAACTTAGAGACGGAAGAAGTT
GAAGAAGATAAAGATTCTAAAAAAAGAAGTTTACTTGCGAATTCAAAGAGACCCACTGCT
AAAAGATCATAGCCATTCAATTAAGATGAAGAGAAAGAAAAATCAGTCACTTAGCCTACC
TCGAATTTAGGAAGCAAAGGTCAATCTTAGTAAGTTGAAAAAGAACAAGCAACAAACTCA
CAAACTTAACAGCCTCAAACAGCACACAGAAGTGGCTCAAGACTCAGCTAGATTCAATCA
AATGCCAATCAAGTTACTTAATAAGCTTAATAGCTATCAAACACCAACAATTCTTCCTCT
ACATCTCTTGAAGTTTCATCAAAAATGCCTAGCTAAATGTCTTAAAAACGCAGACCTATT
GAACTTACTGAAATTTAACAAGGTGATTTCAAGACCGATAATGTCGATAAGATTGAAATC
CAAGGAGATTTTAATGACATAATGACTTCAAGGTTTGAAGTTTTTTGGAAAATCAGATAA
GACAATGTTACCCCTGCTTCCTAAGTCTATTCAGCCTCTTACCTGAGAAGATATGAGCCT
TAAGTTCTCATCGATTTCCTCTTACAACATTCAAACTAATCTCAATTAAGACAAGCTAAT
TAATAACTTACTCATTGA>TTHERM_00125280(gene)
GATGTTTACTGTGAAATCACTCTAATAATTTGTAAATAATTATTTTAAATCCAACCAAAT
ATTTTGTTAATTTTATAAAAATAATAAATTTAAAAATTCTCCTAATTAGTAAGCTAAAAA
AGCAATAAATATTTCAATAAAGAAAATGTCTCAGAAAAAGAGTTTAAAATAAAAGAGAAA
GTAAGACTATAGCTCTGATACTGAAGAGGAAGAAGAGGATCAATATGAAGTTGAAAAGAT
CTTAGACTCTAGATTTAATCCTAAGACCAAGCAAAAAGAATATCTTGTCAAATGGGAAAA
CTGGCCTATTGAAGACTCTACCTGGGAACCTTACGAGCATCTTTCGAATGTCAAAGAAAT
AGTTTAGGCCTTTGAAAAGAAGCAAAAAGCTAACGTTATGCCTCAACCTACTGGACCTAT
CACTAGAGTAAATGCCTAGAAAGACCCTTAAAAAAAGAACAGACTATCTTTGAATTCTTC
TATCAGCAAATCTTTACCTCAAGAAGAAGAAATTTAGACATCTAAAGAAGACAGTAAAAA
ATAAGCAGTCAAAAAATTCCAACCAGCTTCAAGAAGAAAATCTATTAGTTCTTAATCTGA
TTCAGACCTTTAAGCTGAAGAAGTTCCACAGTAGCCTGAAAGTAAAAAAGATAAAAATGA
TGGTGCCTTTGAAGAACCCAACAATGCTGATGCAGACGAAACTGAGCTTGTATTTGAAGA
AATTGTTGACAAAAGAATTCTTGATGGATAAACAGAATATTTAATACGCTTCTAAAATGT
CTCTTAGCCATAATGGGTAGATGTTGGACAATTGATAGCTATTAAAGATGATGTTATAGC
TTATGAAGATAAAATTGCTGCTTAGAGTTAGCTCAATAAGACTAAGAACTTGAAAGAAAA
TGCTAAACATTCATCTTAACAATAGCAAAGCGAAAATAATAACTTAGAGACGGAAGAAGT
TGAAGAAGATAAAGATTCTAAAAAAAGAAGTTTACTTGCGAATTCAAAGAGACCCACTGC
TAAAAGATCATAGCCATTCAATTAAGATGAAGAGAAAGAAAAATCAGTCACTTAGCCTAC
CTCGAATTTAGGAAGCAAAGGTCAATCTTAGTAAGTTGAAAAAGAACAAGCAACAAACTC
ACAAACTTAACAGCCTCAAACAGCACACAGAAGTGGCTCAAGACTCAGCTAGATTCAATC
AAATGCCAATCAAGTTACTTAATAAGCTTAATAGCTATCAAACACCAACAATTCTTCCTC
TACATCTCTTGAAGTTTCATCAAAAATGCCTAGCTAAATGTCTTAAAAACGCAGACCTAT
TGAACTTACTGAAATTTAACAAGGTGATTTCAAGACCGATAATGTCGATAAGATTGAAAT
CCAAGGAGATTTTAATGACATAATGACTTCAAGGTTTGAAGTTTTTTGGAAAATCAGATA
AGACAATGTTACCCCTGCTTCCTAAGTCTATTCAGCCTCTTACCTGAGAAGATATGAGCC
TTAAGTTCTCATCGATTTCCTCTTACAACATTCAAACTAATCTCAATTAAGACAAGCTAA
TTAATAACTTACTCATTGATAAATTATTTAGTGATTACGGCTTGATTAAGCTCTTAAAAC
ATATGATGAACTCAATAAATTTTCTTATTAGAAACCAAAAAAATACTTGGAAAAAAATTT
TTAAAATAAATATATTTCTCATGATAAAAATATATTTTTTTGAAAAATATTGCTGATTAT
TTATTTATTAATTATTACATTTTACATATTTTGTAACGATTGAGTTTTTAGTTTAAAGAG
TTTTAATTTCACATTTT>TTHERM_00125280(protein)
MFTVKSLQQFVNNYFKSNQIFCQFYKNNKFKNSPNQQAKKAINISIKKMSQKKSLKQKRK
QDYSSDTEEEEEDQYEVEKILDSRFNPKTKQKEYLVKWENWPIEDSTWEPYEHLSNVKEI
VQAFEKKQKANVMPQPTGPITRVNAQKDPQKKNRLSLNSSISKSLPQEEEIQTSKEDSKK
QAVKKFQPASRRKSISSQSDSDLQAEEVPQQPESKKDKNDGAFEEPNNADADETELVFEE
IVDKRILDGQTEYLIRFQNVSQPQWVDVGQLIAIKDDVIAYEDKIAAQSQLNKTKNLKEN
AKHSSQQQQSENNNLETEEVEEDKDSKKRSLLANSKRPTAKRSQPFNQDEEKEKSVTQPT
SNLGSKGQSQQVEKEQATNSQTQQPQTAHRSGSRLSQIQSNANQVTQQAQQLSNTNNSSS
TSLEVSSKMPSQMSQKRRPIELTEIQQGDFKTDNVDKIEIQGDFNDIMTSRFEVFWKIRQ
DNVTPASQVYSASYLRRYEPQVLIDFLLQHSNQSQLRQANQQLTH