Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00189180Standard Name
HHT2 (Histone H Three)Aliases
PreTt08668 | 15.m00379 | 3697.m00091Description
HHT2 predicted protein; Histone H3; one of the four histones (H2A, H2B, H3 and H4) that comprise the protein core of the eukaryotic nucleosome; lysine-9 methylated in heterochromatin; encoded protein identical to Hht1p; upregulated in HHT3 knockouts; Histone H3/CENP-AGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
No Data fetched for Gene Ontology Annotations
 Domains
Domains
- ( PF00125 ) Core histone H2A/H2B/H3/H4
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
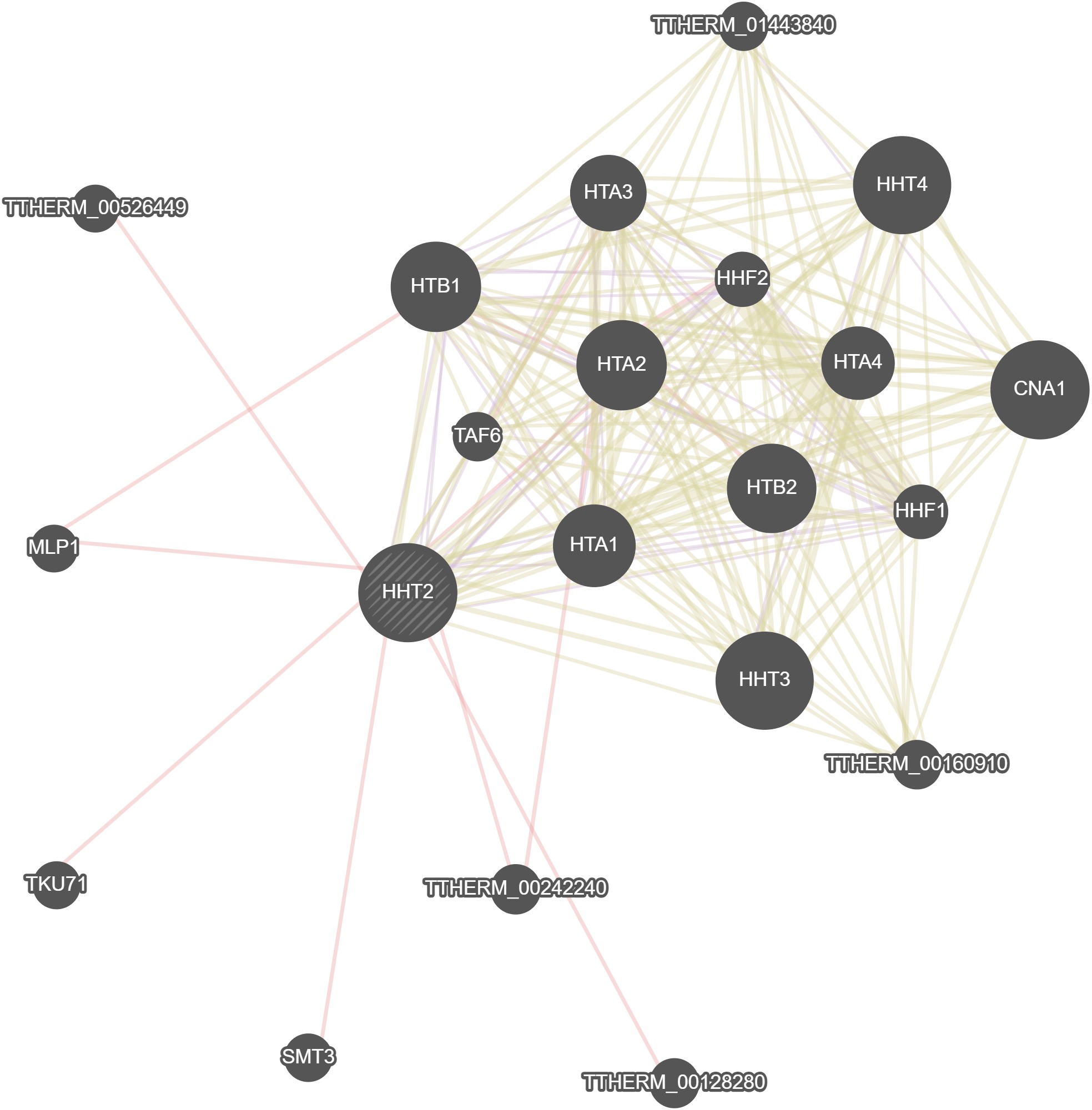
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
- ( SD01317 ) Micronucleus: Neo2 into HHT1 and KO HHT2/HHF2 gene and its intergenic region; KO of HHF1
- ( SD01361 ) Micronucleus: Neo KO of all major HHT genes and HHF genes
- ( SD01362 ) Micronucleus: Neo KO of all major HHT genes and HHF genes
- ( SD01370 ) Macronucleus: Neo2 cassettes KO HHT2
- ( SD01371 ) Macronucleus: c-terminal HHT2 FLAG tagged
- ( SD01373 ) Macronucleus: hht2[3’ neo2,FLAGc]
- ( SD01778 ) Micronucleus: Neo KO of HHT3, Bsr KO of HHT4, GFP tagged HHT2 Macronucleus: Neo KO of HHT3, Bsr KO of HHT4, GFP tagged HHT2
- ( SD01852 ) Micronucleus: c-terminal GFP tagged HHT2,with 3’ flanking neo2
- ( SD01853 ) Micronucleus: c-terminal GFP tagged HHT2,with 3’ flanking neo2
- ( SD01864 ) Micronucleus: Neo3 KO of major H3s and HHT3
- ( SD01865 ) Micronucleus: Neo3 KO of major H3s and HHT3
- ( SD01866 ) Micronucleus: Neo3 KO of major H3s and HHT3
- ( SD01869 ) Micronucleus: Neo2 KO of HHT1 and HHT2/HHF2
- ( SD01870 ) Micronucleus: Neo2 KO of HHT1 and HHT2/HHF2
- ( SD01871 ) Micronucleus: Neo2 KO of HHT1 and HHT2/HHF2
- ( SD01872 ) Micronucleus: Neo2 KO of HHT1 and HHT2/HHF2
- ( SD01880 ) Micronucleus: Neo2 KO of HHT3 and HHT2/HHF2 gene with its intergenic region
- ( SD01881 ) Micronucleus: Neo3 KO of HHT1,HHT2/HHF2
- ( SD01886 ) Macronucleus: KO of HHT1, HHT2 has mutation S10A
- ( SD01887 ) Macronucleus: KO of HHT1, HHT2 has mutation S10A
- ( SD01888 ) Macronucleus: KO of HHT1, HHT2 has mutation S10A
- ( SD01889 ) Macronucleus: KO of HHT1, HHT2 has mutation S10A
- ( SD01893 ) Macronucleus: HHT1 replaced by neo2, HHT2 has S10A mutation
- ( SD01894 ) Macronucleus: HHT1 replaced by neo2, HHT2 has S10A mutation
- ( SD01898 ) Macronucleus: HHT2 with GFP c-terminal tag replaces the HHT3 coding region
- ( SD01899 ) Macronucleus: HHT2 with GFP c-terminal tag replaces the HHT3 coding region
- ( SD01949 ) Micronucleus: Neo2 ko of HHT1 and HHT2/HHF2
- ( SD01955 ) Micronucleus: Neo2 ko of HHT1 and HHT2/HHF2
- ( SD01956 ) Micronucleus: Neo2 ko of HHT1 and HHT2/HHF2
- ( SD01957 ) Micronucleus: Neo2 ko of HHT1 and HHT2/HHF2
- ( SD01958 ) Micronucleus: Neo2 ko of HHT1 and HHT2/HHF2
- ( SD01959 ) Micronucleus: Neo2 ko of HHT1 and HHT2/HHF2
- ( SD01960 ) Micronucleus: Neo2 ko of HHT1 and HHT2/HHF2
- ( SD01994 ) Micronucleus: Neo3 KO of HHT!,HHT2/HHF2,HHT3
- ( SD01995 ) Micronucleus: Neo3 KO of HHT!,HHT2/HHF2,HHT3
- ( SD02005 )
- ( SD02006 )
- ( SD02036 ) Micronucleus: Neo into HHT1, HHF1, HHT2/HHF2
- ( SD02037 ) Micronucleus: Neo into HHT1, HHF1, HHT2/HHF2
- ( SD02038 ) Micronucleus: Neo into HHT1, HHF1, HHT2/HHF2
- ( SD02039 ) Micronucleus: Neo into HHT1, HHF1, HHT2/HHF2
- ( SD02040 ) Micronucleus: Neo into HHT1, HHF1, HHT2/HHF2
- ( SD02041 ) Micronucleus: Neo into HHT1, HHF1, HHT2/HHF2
- ( SD02042 ) Micronucleus: Neo KO of all major HHT genes and HHF genes Macronucleus: WT HHF2 and mutated HHT2 with K9Q
- ( SD02148 ) Micronucleus: Neo2 KO of HHT1 and HHT2/HHF2
- ( SD02159 ) Macronucleus: HHT1 replaced by neo2, HHT2 has S10A, S28A mutations
- ( SD02160 ) Macronucleus: HHT1 replaced by neo2, HHT2 has S10A, S28A mutations
- ( SD02161 ) Macronucleus: HHT1 replaced by neo2, HHT2 has S10A, S28A mutations
- ( SD02162 ) Macronucleus: HHT1 replaced by neo2, HHT2 has S10A, S28A mutations
- ( SD02304 ) Micronucleus: Neo3 KO of HHT!,HHT2/HHF2,HHT3
- ( SD02310 ) Micronucleus: Neo KO of all major HHT genes and HHF genes Macronucleus: WT HHF2 and mutated HHT2 with K27R
- ( SD02311 ) Micronucleus: Neo KO of all major HHT genes and HHF genes Macronucleus: WT HHF2 and mutated HHT2 with K9Q
- ( SD02312 ) Macronucleus: c-terminal HHT2 FLAG tagged
- ( SD02313 ) Macronucleus: c-terminal HHT2 FLAG tagged
- ( SD02320 ) Micronucleus: Neo KO of all major HHT genes and HHF genes Macronucleus: WT HHF2 and mutated HHT2 with K9Q
- ( SD02373 ) Micronucleus: Neo knockout of HHT1, HHT2, HHF1 Macronucleus: KO of HHT1, HHF1;and for HHT2 have mutations S10A and S28A
- ( SD02374 ) Micronucleus: Neo knockout of HHT1,HHT2, HHF1 Macronucleus: KO of HHT1, HHF1; and for HHT2 have mutations S10A and S28A
- ( SD02375 ) Micronucleus: Neo knockout of HHT1,HHT2, HHF1 Macronucleus: KO of HHT1, HHF1;and for HHT2 have mutations S10A and S28A
- ( SD02376 ) Micronucleus: Neo knockout of HHT1,HHT2, HHF1 Macronucleus: KO of HHT1, HHF1;and for HHT2 have mutations S10A and S28A
- ( SD02377 ) Micronucleus: Neo knockout of HHT1,HHT2, HHF2 Macronucleus: KO of HHT1, HHF2; and for HHT2 have mutation S28A
- ( SD02378 ) Micronucleus: Neo knockout of HHT1,HHT2, HHF2 Macronucleus: KO of HHT1, HHF2; and for HHT2 have mutation S28A
- ( SD02379 ) Micronucleus: Neo knockout of HHT1,HHT2, HHF2 Macronucleus: KO of HHT1, HHF2; and for HHT2 have mutation S28A
- ( SD02380 ) Micronucleus: Neo knockout of HHT1,HHT2, HHF2 Macronucleus: KO of HHT1, HHF2; and for HHT2 have mutation S28A
- ( SD02526 ) Micronucleus: Neo2 in 3’ flank and S10E mutation in HHT2
- ( SD02527 ) Micronucleus: Neo KO of HHT1. HHT3 and HHT2 have S10E mutation and neo2 in 3’ flanking region. Macronucleus: Neo KO of HHT1. HHT3 and HHT2 have S10E mutation and neo2 in 3’ flanking region
- ( SD02528 ) Micronucleus: Neo KO of HHT1. HHT3 and HHT2 have S10E mutation and neo2 in 3’ flanking region. Macronucleus: Neo KO of HHT1. HHT3 and HHT2 have S10E mutation and neo2 in 3’ flanking region
- ( SD02544 ) Micronucleus: Neo 2 KO of HHT1 and of HHT2/HHF2 Macronucleus: Neo 2 Ko of HHT1, WT HHT2/HHF2
- ( SD02545 ) Micronucleus: Neo 2 KO of HHT1 and of HHT2/HHF2 Macronucleus: Neo 2 Ko of HHT1, WT HHT2/HHF2
- ( SD02546 ) Micronucleus: KO of HHT1,HHT2,HHF2 Macronucleus: HHF1 ko and HHF2/HHT2 is now WT
- ( SD02547 ) Micronucleus: KO of HHT1,HHT2,HHF2 Macronucleus: HHF1 ko and HHF2/HHT2 is now WT
- ( SD02614 ) Micronucleus: Neo KO of HHT3, Bsr KO of HHT4, GFP tagged HHT2 Macronucleus: Neo KO of HHT3, Bsr KO of HHT4, GFP tagged HHT2
- ( SD02623 ) Micronucleus: Neo2 replaces the HHF2 gene and HHT2 gene and the sequence between them
- ( SD02624 ) Micronucleus: Neo2 replaces the HHF2 gene and HHT2 gene and the sequence between them
- ( SD02625 ) Micronucleus: Neo2 replaces the HHF2 gene and HHT2 gene and the sequence between them
- ( SD02719 ) Macronucleus: HHT2 with GFP c-terminal tag replaces the MTT1 coding region
- ( SD02791 ) Micronucleus: GFP tagged HHT2 Macronucleus: GFP tagged HHT2
- ( SD02792 ) Macronucleus: Neo2 cassettes KO HHT2
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2209 PDD1, HHT2, DCL1, PDD3, TWI1 A proposed model for the mechanism of programmed DNA elimination in Tetrahymena is based on the timing of expression, cellular distribution, mutant phenotypes, and predicted functions of the protein and RNA components involved. In this model, both strands of the micronuclear genome (or perhaps only the portions containing internal eliminated sequences) are transcribed early in conjugation to produce large non-genic, double-stranded RNAs. This transcription is likely performed by RNA Polymerase II, based on the localization of its subunit Rpb3p to the micronucleus during this time. These transcripts pass to the cytoplasm where they are processed into short (~28 nucleotide) scan RNAs (scnRNA) by the dicer-like protein Dcl1p, similar to the production of the small inhibitory RNAs (siRNA) central to the RNA interference (RNAi) pathway of other eukaryotes. The scnRNAs complex with Twi1p, a member of the PPD (PAZ and Piwi Domain) protein family, whose members are commonly involved in RNAi and related processes. The scnRNA/Twi1p complexes enter the old macronucleus, where scnRNAs homologous to DNA sequences found there are degraded. The remaining scnRNAs, comprised of micronuclear-restricted sequences, are transferred to the developing macronucleus. There, histone H3 proteins (Hht1p, Hht2p) that are bound to sections of the genome sharing identity to the scnRNAs are methylated on lysine-9. This modification, which is often associated with the formation of heterochromatin, is recognized by one or more of the chromodomains belonging to Pdd1p and Pdd3p. Regions of DNA associated with these modified histones are eliminated from the developing macronuclear genome.
 Associated Literature
Associated Literature
- Ref:16908532: Cui B, Liu Y, Gorovsky MA (2006) Deposition and function of histone H3 variants in Tetrahymena thermophila. Molecular and cellular biology 26(20):7719-30
- Ref:15701804: Liu Y, Song X, Gorovsky MA, Karrer KM (2005) Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryotic cell 4(2):421-31
- Ref:10199406: Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD (1999) Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97(1):99-109
- Ref:9636175: Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD (1998) Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America 95(13):7480-4
 Sequences
Sequences
>TTHERM_00189180(coding)
ATGGCTAGAACTAAATAAACTGCTAGAAAGTCCACTGGTGCTAAGGCCCCCAGAAAATAA
CTCGCTTCCAAGGCCGCCAGAAAGTCTGCCCCCGCCACTGGTGGTATCAAGAAGCCCCAC
AGATTCAGACCTGGTACCGTCGCTCTCAGAGAAATCAGAAAGTACCAAAAGTCCACTGAT
TTGTTGATCAGAAAGCTCCCCTTCTAAAGATTGGTCAGAGATATTGCTCACGAATTCAAG
GCTGAACTCAGATTCTAATCTTCTGCCGTTCTTGCTCTCCAAGAAGCTGCTGAAGCTTAC
CTCGTCGGTCTCTTCGAAGATACCAACTTGTGCGCTATCCACGCTAGAAGAGTTACTATT
ATGACCAAGGATATGCAACTCGCTAGAAGAATTAGAGGTGAAAGATTCTGA>TTHERM_00189180(gene)
AAATAATAACCTTCAAAAGAAAAGTCAAAAAGACAATCCACTATAAATACATAAGCAAAA
ATGGCTAGAACTAAATAAACTGCTAGAAAGTCCACTGGTGCTAAGGCCCCCAGAAAATAA
CTCGCTTCCAAGGCCGCCAGAAAGTCTGCCCCCGCCACTGGTGGTATCAAGAAGCCCCAC
AGATTCAGACCTGGTACCGTCGCTCTCAGAGAAATCAGAAAGTACCAAAAGTCCACTGAT
TTGTTGATCAGAAAGCTCCCCTTCTAAAGATTGGTCAGAGATATTGCTCACGAATTCAAG
GCTGAACTCAGATTCTAATCTTCTGCCGTTCTTGCTCTCCAAGAAGCTGCTGAAGCTTAC
CTCGTCGGTCTCTTCGAAGATACCAACTTGTGCGCTATCCACGCTAGAAGAGTTACTATT
ATGACCAAGGATATGCAACTCGCTAGAAGAATTAGAGGTGAAAGATTCTGAGCATAATAT
AACAACTAGTCTCTAAATAATCAACATACCACATATATAATCATTACTTAATTTATCAAA
ATATCATATGTATATTATAACTTTCGTGTGAGTACATAAAACCTTTTTTAGAGTTTTCGA
GTTTTCAAAAAAGATAATCCAAAATAACTTCCTCCATATATATAAATATGCATATATATA
AAAATAATTATTAAATTCCACAATATAAAATTTATTTGTTTTTTTATTCAATATCATTCA
TTTGTATT>TTHERM_00189180(protein)
MARTKQTARKSTGAKAPRKQLASKAARKSAPATGGIKKPHRFRPGTVALREIRKYQKSTD
LLIRKLPFQRLVRDIAHEFKAELRFQSSAVLALQEAAEAYLVGLFEDTNLCAIHARRVTI
MTKDMQLARRIRGERF