Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01207660Standard Name
CDC2 (Cell Division Cycle 2)Aliases
PreTt09119 | 274.m00036Description
CDC2 cyclin-dependent kinase-like Serine/Threonine kinase family protein; Homolog of the catalytic subunit of the major cell cycle cyclin-dependent kinase (cdk); studies suggest cdc2p phosphorylates HHO1; expression positively regulated by HHO1 phosphorylation during starvation, resulting in a positive feedback; Cyclin-dependent kinaseGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- cyclin-dependent protein kinase holoenzyme complex (ISS) | GO:0000307
Molecular Function
- cyclin-dependent protein serine/threonine kinase activity (ISS) | GO:0004693
Biological Process
- cell cycle (ISS) | GO:0007049
- obsolete histone phosphorylation (IGI) | GO:0016572
 Domains
Domains
- ( PF00069 ) Protein kinase domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
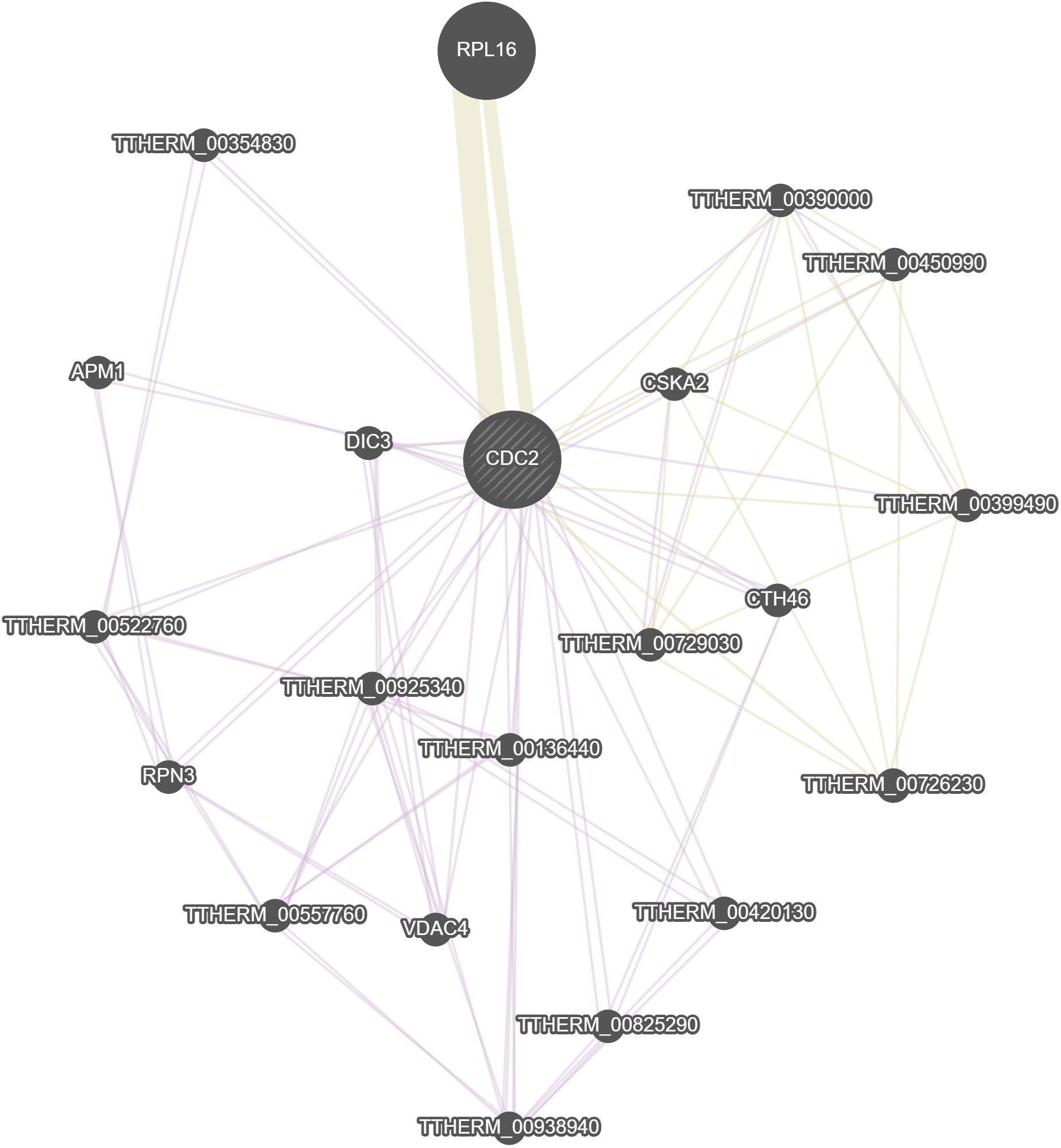
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
- ( SD02572 ) Macronucleus: CDC2 gene has the 942 CDC2 5’ flanking region that includes the H1 dephosphorylation enriched region and GFP coding replacing CDC2 coding. Also neo3 in 3’ flanking. region.
- ( SD02573 ) Macronucleus: CDC2 gene has the 1.6 kb just upstream of ATG lacking and GFP coding replacing CDC2 coding. Also neo3 in 3’ flanking. region.
- ( SD02574 ) Macronucleus: CDC2 gene has 5’ flanking sequence of 280 bp and GFP coding replacing CDC2 coding. Also neo3 in 3’ flanking.
- ( SD02575 ) Macronucleus: CDC2 gene has 5’ flanking sequence of 247 bp and GFP coding replacing CDC2 coding. Also neo3 in 3’ flanking.
- ( SD02577 ) Macronucleus: CDC2 gene has the 942 CDC2 5’ flanking region that includes the H1 dephosphorylation enriched region and GFP coding replacing CDC2 coding. Also neo3 in 3’ flanking. region.
- ( SD02578 ) Macronucleus: CDC2 gene 5’ flanking sequence of dephosphorylated HI enriched region and coding removed and replaced with GFP gene. Also neo3 in 3’ flanking.
- ( SD02822 ) Macronucleus: BTU1 locus replaced by far 5’ BTU1 5’--neo3--CDC2 5’ flanking 1.5 kb immediately upstream of ATG--GFP coding--BTU1 3’
- ( SD02823 ) Macronucleus: BTU1 locus replaced by far 5’ BTU1 5’--neo3--CDC2 5’ flanking 946 bp immediately upstream of ATG--GFP coding--BTU1 3’
- ( SD02824 ) Macronucleus: BTU1 locus replaced by far 5’ BTU1 5’--neo3--CDC2 5’ flanking 480bp immediately upstream of ATG--GFP coding--BTU1 3’
- ( SD02825 ) Macronucleus: BTU1 locus replaced by far 5’ BTU1 5’--neo3--CDC2 5’ flanking 480 bp immediately upstream of ATG--GFP coding--BTU1 3’
- ( SD02826 ) Macronucleus: BTU1 locus replaced by far 5’ BTU1 5’--neo3--CDC2 5’ flanking 480 bp immediately upstream of ATG--GFP coding--BTU1 3’
- ( SD02827 ) Macronucleus: BTU1 locus replaced by far 5’ BTU1 5’--neo3--CDC2 5’ flanking 480 bp immediately upstream of ATG--GFP coding--BTU1 3’
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2216 NGOA1, HHO1, CYP1, CDC2 HHO1 knockouts show no global increase or decrease in the amount of transcription in the cell; however, these same knockouts also show that Hho1p is important for the transcriptional regulation of individual genes in response to stimuli, such as starvation. The differential regulation of Hho1p by phosphorylation under vegetative growth and starvation conditions has been well studied. During vegetative growth, Hho1p is phosphorylated on five closely spaced residues, preventing it from interacting with chromatin, likely by interfering with its ability to bind DNA. Under these conditions, expression is increased for CDC2, a homolog of the cyclin dependent kinases responsible for histone H1 phosphorylation, possibly creating a positive feedback loop that promotes the cell cycle. During starvation conditions, Hho1p is dephosphorylated, allowing it to bind to chromatin. This stimulates the expression of some genes, including ngoA, and protease genes such as CYP1, while inhibiting expression of other genes, such as CDC2. This decrease in CDC2 expression may be responsible for cell cycle arrest during starvation.
 Associated Literature
Associated Literature
- Ref:21562224: Vonderfecht T, Stemm-Wolf AJ, Hendershott M, Giddings TH, Meehl JB, Winey M (2011) The two domains of centrin have distinct basal body functions in Tetrahymena. Molecular biology of the cell 22(13):2221-34
- Ref:17194754: Song X, Gorovsky MA (2007) Unphosphorylated H1 is enriched in a specific region of the promoter when CDC2 is down-regulated during starvation. Molecular and cellular biology 27(5):1925-33
- Ref:17715364: Parker K, Maxson J, Mooney A, Wiley EA (2007) Class I histone deacetylase Thd1p promotes global chromatin condensation in Tetrahymena thermophila. Eukaryotic cell 6(10):1913-24
- Ref:16933976: Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA,
- Ref:15870266: Dou Y, Song X, Liu Y, Gorovsky MA (2005) The H1 phosphorylation state regulates expression of CDC2 and other genes in response to starvation in Tetrahymena thermophila. Molecular and cellular biology 25(10):3914-22
- Ref:12832485: Mohammad M, York RD, Hommel J, Kapler GM (2003) Characterization of a novel origin recognition complex-like complex: implications for DNA recognition, cell cycle control, and locus-specific gene amplification. Molecular and cellular biology 23(14):5005-17
- Ref:12183062: Zhang H, Huang X, Tang L, Zhang QJ, Frankel J, Berger JD (2002) A cyclin-dependent protein kinase homologue associated with the basal body domains in the ciliate Tetrahymena thermophila. Biochimica et biophysica acta 1591(1-3):119-128
- Ref:11456317: Sugii M, Fujishima M (2001) Purification of GVBD-inducing protein from the ciliate Tetrahymena thermophila. The Journal of eukaryotic microbiology 48(4):414-24
- Ref:10975520: De Souza CP, Osmani AH, Wu LP, Spotts JL, Osmani SA (2000) Mitotic histone H3 phosphorylation by the NIMA kinase in Aspergillus nidulans. Cell 102(3):293-302
- Ref:10527850: Gonda K, Nishibori K, Ohba H, Watanabe A, Numata O (1999) Molecular cloning of the gene for p85 that regulates the initiation of cytokinesis in Tetrahymena. Biochemical and biophysical research communications 264(1):112-8
- Ref:10329641: Mizzen CA, Dou Y, Liu Y, Cook RG, Gorovsky MA, Allis CD (1999) Identification and mutation of phosphorylation sites in a linker histone. Phosphorylation of macronuclear H1 is not essential for viability in tetrahymena. The Journal of biological chemistry 274(21):14533-6
- Ref:9819444: Won KA, Schumacher RJ, Farr GW, Horwich AL, Reed SI (1998) Maturation of human cyclin E requires the function of eukaryotic chaperonin CCT. Molecular and cellular biology 18(12):7584-9
- Ref:9811850: Huang H, Wiley EA, Lending CR, Allis CD (1998) An HP1-like protein is missing from transcriptionally silent micronuclei of Tetrahymena. Proceedings of the National Academy of Sciences of the United States of America 95(23):13624-9
- Ref:8995382: Sweet MT, Carlson G, Cook RG, Nelson D, Allis CD (1997) Phosphorylation of linker histones by a protein kinase A-like activity in mitotic nuclei. The Journal of biological chemistry 272(2):916-23
- Ref:8264578: Wu M, Allis CD, Sweet MT, Cook RG, Thatcher TH, Gorovsky MA (1994) Four distinct and unusual linker proteins in a mitotically dividing nucleus are derived from a 71-kilodalton polyprotein, lack p34cdc2 sites, and contain protein kinase A sites. Molecular and cellular biology 14(1):10-20
- Ref:1453357: Fujishima M, Katsu Y, Ogawa E, Sakimura M, Yamashita M, Nagahama Y (1993) Meiosis-reinitiation-inducing factor of Tetrahymena functions upstream of M-phase-promoting factor. The Journal of protozoology 39(6):683-90
- Ref:1297353: Masui Y (1993) Towards understanding the control of the division cycle in animal cells. Biochemistry and cell biology = Biochimie et biologie cellulaire 70(10-11):920-45
- Ref:8306826: Sweet MT, Allis CD (1993) Phosphorylation of linker histones by cAMP-dependent protein kinase in mitotic micronuclei of Tetrahymena. Chromosoma 102(9):637-47
- Ref:2065655: Roth SY, Collini MP, Draetta G, Beach D, Allis CD (1991) A cdc2-like kinase phosphorylates histone H1 in the amitotic macronucleus of Tetrahymena. The EMBO journal 10(8):2069-75
- Ref:17821280: Harrison JA, Fowler EH (1945) AN ANTIGEN-ANTIBODY REACTION WITH TETRAHYMENA WHICH RESULTS IN DYSTOMY. Science (New York, N.Y.) 102(2638):65-6
 Sequences
Sequences
>TTHERM_01207660(coding)
ATGGTTATGAAAAATAGCAAGCTTGAGCGTTACGAAAAGCTCAATAAGCTCGGTGAAGGT
ACATATGGTGTTGTATATAAAGCAAGAGAAAAAACTACCAAAGAATTGTATGCCTTGAAA
AAAATCCGTTTAGAAAGTGAAGATGAAGGTATTCCCTCTACCGCAATCAGAGAAATCTCT
CTCTTAAAGGAGTTATAACATCCTAATGTTGTTAGATTGCACGATGTCATCCATTCAAAT
AAGAAACTCGTTTTAGTTTTTGAATTCGTTGATTAAGATTTAAAAAAATTCATGAATAAC
TTCAAGGATAAAGGACTCGATCCTCATATTATTAAGAGCTTACTTTATCAATTATTGAAG
GGTATTGAAGTTTGTCATAAAAATAAAATTCTTCATAGAGATTTGAAGCCTCAAAATTTG
CTAATTTCAAAGGAATGTATCCTTAAGTTAGCAGATTTCGGTTTGGCTAGAGCTTCTGGT
ATTCCTGTAAAGAATTATACACATGAAGTCGTTACTTTATGGTACAGACCTCCTGATGTC
CTTTTGGGATCTAAGCACTATTCCACTTCAATTGATATCTGGAGTATTGGTTGTATATTT
GCTGAAATGGTTAACTTAAAACCCTTGTTCCCTGGTAATTCAGAAACAGACGAATTAAAG
CGTATCTTTAAGTTGACAGGTACTCCATGTGTTGAAAAATGGCCTGGGCTTGCCGATCTT
CCTAATTGGAAGGCTGATGCATTTGAAAAATATCCTGGAGAGCCTCTTCAAAATATTTGC
CCCAAGCTTGATGAACTCGGTCTCGACTTACTTGGTAAAATGTTAAGATGCAACCCATAA
GAAAGAATTACTGCCAAAGCAGGTTTAGAACATCCCTATTTCAACGATATTCCTGACACA
TTAAAGAAGTTATATCAATCAAACTGA>TTHERM_01207660(gene)
TAATAATTAAATAAAAAAATTAAACTAATAAAAAGCAAATAAATAATCTGACAGTAAAAA
TGGTTATGAAAAATAGCAAGCTTGAGCGTTACGAAAAGCTCAATAAGCTCGGTGAAGGTA
CATATGGTGTTGTATATAAAGCAAGAGAAAAAACTACCAAAGAATTGTATGCCTTGAAAA
AAATCCGTTTAGAAAGTGAAGATGAAGGTATTCCCTCTACCGCAATCAGAGAAATCTCTC
TCTTAAAGGAGTTATAACATCCTAATGTTGTTAGATTGCACGATGTCATCCATTCAAATA
AGAAACTCGTTTTAGTTTTTGAATTCGTTGATTAAGATTTAAAAAAATTCATGAATAACT
TCAAGGATAAAGGACTCGATCCTCATATTATTAAGAGCTTACTTTATCAATTATTGAAGG
GTATTGAAGTTTGTCATAAAAATAAAATTCTTCATAGAGATTTGAAGCCTCAAAATTTGC
TAATTTCAAAGGAATGTATCCTTAAGTTAGCAGATTTCGGTTTGGCTAGAGCTTCTGGTA
TTCCTGTAAAGAATTATACACATGAAGTCGTTACTTTATGGTACAGACCTCCTGATGTCC
TTTTGGGATCTAAGCACTATTCCACTTCAATTGATATCTGGAGTATTGGTTGTATATTTG
CTGAAATGGTTAACTTAAAACCCTTGTTCCCTGGTAATTCAGAAACAGACGAATTAAAGC
GTATCTTTAAGTTGACAGGTACTCCATGTGTTGAAAAATGGCCTGGGCTTGCCGATCTTC
CTAATTGGAAGGCTGATGCATTTGAAAAATATCCTGGAGAGCCTCTTCAAAATATTTGCC
CCAAGCTTGATGAACTCGGTCTCGACTTACTTGGTAAAATGTTAAGATGCAACCCATAAG
AAAGAATTACTGCCAAAGCAGGTTTAGAACATCCCTATTTCAACGATATTCCTGACACAT
TAAAGAAGTTATATCAATCAAACTGATAAATATGAATAAGCTCGAAGGTAGCTTGAAAAT
GTTTTAAAAATTAAAATTAAATCCTTCGATAAATAGTAAAATAAATAAAATATTATGAAC
AAATTAAAAAAATGTAAGCAATATTATTTGATAAATAAGCTTTAATAAACTAACTATTTC
TCCATTTGTAAATGTAGTTAAAATATATTTTTTGTAACTTTAATAGCAGATTTATTATTT
TTCTAACTCTTTTT>TTHERM_01207660(protein)
MVMKNSKLERYEKLNKLGEGTYGVVYKAREKTTKELYALKKIRLESEDEGIPSTAIREIS
LLKELQHPNVVRLHDVIHSNKKLVLVFEFVDQDLKKFMNNFKDKGLDPHIIKSLLYQLLK
GIEVCHKNKILHRDLKPQNLLISKECILKLADFGLARASGIPVKNYTHEVVTLWYRPPDV
LLGSKHYSTSIDIWSIGCIFAEMVNLKPLFPGNSETDELKRIFKLTGTPCVEKWPGLADL
PNWKADAFEKYPGEPLQNICPKLDELGLDLLGKMLRCNPQERITAKAGLEHPYFNDIPDT
LKKLYQSN