Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_01337400Standard Name
HPL2 (Heterochromatin Protein 1 Like 2)Aliases
PreTt02558 | 333.m00018 | 3795.m00073 | TCD1 | JUB5Description
HPL2 heterochromatin protein; Heterochromatin protein 1; Chromo domain-containing protein LHP1Genome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Cellular Component
- brahma complex (IEA) | GO:0035060
- cellulosome (IEA) | GO:0043263
- ionotropic glutamate receptor complex (IEA) | GO:0008328
- obsolete ectoplasm (IEA) | GO:0043265
- obsolete integral component of mitochondrial membrane (IEA) | GO:0032592
- subrhabdomeral cisterna (IEA) | GO:0016029
Molecular Function
- 11-deoxycorticosterone binding (IEA) | GO:1903878
- 17alpha-hydroxyprogesterone binding (IEA) | GO:1903880
- 2-keto-3-deoxy-L-rhamnonate aldolase activity (IEA) | GO:0106099
- 3-beta-hydroxy-delta5-steroid dehydrogenase (NAD+) activity (IEA) | GO:0003854
- 3-hydroxybutyrate dehydrogenase activity (IEA) | GO:0003858
- 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity (IEA) | GO:0008413
- acylglycerone-phosphate reductase (NADP+) activity (IEA) | GO:0000140
- aminotriazole transporter activity (IEA) | GO:0015241
- autotransporter activity (IEA) | GO:0015474
- beta-pinacene synthase activity (IEA) | GO:0106100
- complement component C5a receptor activity (IEA) | GO:0004878
- delta-type opioid receptor binding (IEA) | GO:0031850
- dicarboxylate:phosphate antiporter activity (IEA) | GO:0015364
- dolichyl pyrophosphate Glc2Man9GlcNAc2 alpha-1,2-glucosyltransferase activity (IEA) | GO:0106073
- ferredoxin-NAD(P) reductase activity (IEA) | GO:0008937
- ferric enterobactin:proton symporter activity (IEA) | GO:0015345
- ferric triacetylfusarinine C:proton symporter activity (IEA) | GO:0015346
- FMN reductase (NADH) activity (IEA) | GO:0052874
- high-affinity ferric iron transmembrane transporter activity (IEA) | GO:0015092
- juvenile hormone receptor activity (IEA) | GO:0004880
- L-gamma-glutamyl-L-propargylglycine hydroxylase activity (IEA) | GO:0062148
- lanosterol synthase activity (IEA) | GO:0000250
- lipopolysaccharide heptosyltransferase activity (IEA) | GO:0008920
- lipopolysaccharide-1,6-galactosyltransferase activity (IEA) | GO:0008921
- mannitol-1-phosphate 5-dehydrogenase activity (IEA) | GO:0008926
- mercury ion transmembrane transporter activity (IEA) | GO:0015097
- metarhodopsin binding (IEA) | GO:0016030
- microtubule severing ATPase activity (IEA) | GO:0008568
- mitochondrial single subunit type RNA polymerase activity (IEA) | GO:0001065
- obsolete calcium-dependent secreted phospholipase A2 activity (IEA) | GO:0004625
- obsolete dicarboxylate (succinate/fumarate/malate) antiporter activity (IEA) | GO:0015363
- obsolete hydroxy/aromatic amino acid permease activity (IEA) | GO:0015507
- obsolete phosphatidylinositol 3-kinase, class I, catalyst activity (IEA) | GO:0015072
- obsolete sodium-transporting two-sector ATPase activity (IEA) | GO:0015443
- obsolete tocopherol omega-hydroxylase activity (IEA) | GO:0052870
- obsolete zinc, iron permease activity (IEA) | GO:0015342
- oxalyl-CoA decarboxylase activity (IEA) | GO:0008949
- phosphatidylserine decarboxylase activity (IEA) | GO:0004609
- phosphoenolpyruvate carboxykinase (ATP) activity (IEA) | GO:0004612
- phosphoenolpyruvate carboxykinase activity (IEA) | GO:0004611
- phosphoglycerate dehydrogenase activity (IEA) | GO:0004617
- plastid single subunit type RNA polymerase activity (IEA) | GO:0001066
- proteinase activated receptor binding (IEA) | GO:0031871
- siderophore uptake transmembrane transporter activity (IEA) | GO:0015344
- siderophore-iron transmembrane transporter activity (IEA) | GO:0015343
- sphingomyelin phosphodiesterase activity (IEA) | GO:0004767
- stretch-activated, monoatomic cation-selective, calcium channel activity (IEA) | GO:0015275
- Swi5-Sfr1 complex binding (IEA) | GO:1905334
- transcription regulatory region nucleic acid binding (IEA) | GO:0001067
- type 4 neuropeptide Y receptor binding (IEA) | GO:0031844
- undecaprenol kinase activity (IEA) | GO:0009038
- zinc efflux active transmembrane transporter activity (IEA) | GO:0015341
Biological Process
- 3-keto-sphinganine metabolic process (IEA) | GO:0006666
- cadmium ion transport (IEA) | GO:0015691
- cell wall modification involved in fruit ripening (IEA) | GO:0009829
- cellular response to diacyl bacterial lipopeptide (IEA) | GO:0071726
- cellular response to homocysteine (IEA) | GO:1905375
- coenzyme A biosynthetic process (IEA) | GO:0015937
- double-stranded RNA biosynthetic process (IEA) | GO:0062103
- endoplasmic reticulum to cytosol transport (IEA) | GO:1903513
- epithelium development (IEA) | GO:0060429
- galactose to glucose-1-phosphate metabolic process (IEA) | GO:0061612
- glycolytic process from glycerol (IEA) | GO:0061613
- glycolytic process through fructose-6-phosphate (IEA) | GO:0061615
- heme A biosynthetic process (IEA) | GO:0006784
- heme biosynthetic process (IEA) | GO:0006783
- immunoglobulin heavy chain V-D-J recombination (IEA) | GO:0071707
- inactivation of recombination (HML) (IEA) | GO:0007537
- induction of apoptosis by extracellular signals (IEA) | GO:0008624
- interleukin-12 production (IEA) | GO:0032615
- interleukin-16 production (IEA) | GO:0032619
- lipid homeostasis (IEA) | GO:0055088
- lung connective tissue development (IEA) | GO:0060427
- lung epithelium development (IEA) | GO:0060428
- lung pattern specification process (IEA) | GO:0060432
- lung saccule development (IEA) | GO:0060430
- maltoheptaose catabolic process (IEA) | GO:2001123
- maltoheptaose metabolic process (IEA) | GO:2001122
- mannotriose transport (IEA) | GO:2001095
- metanephric mesenchyme morphogenesis (IEA) | GO:0072133
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902647
- negative regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902642
- negative regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002599
- negative regulation of cellular glucuronidation (IEA) | GO:2001030
- negative regulation of chemokine (C-C motif) ligand 1 production (IEA) | GO:0071653
- negative regulation of complement-dependent cytotoxicity (IEA) | GO:1903660
- negative regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001033
- negative regulation of L-glutamine biosynthetic process (IEA) | GO:0062133
- negative regulation of PERK-mediated unfolded protein response (IEA) | GO:1903898
- negative regulation of phytoalexin metabolic process (IEA) | GO:0052321
- negative regulation of protein localization to presynapse (IEA) | GO:1905385
- negative regulation of somitomeric trunk muscle development (IEA) | GO:0014710
- neutral lipid metabolic process (IEA) | GO:0006638
- obsolete age dependent accumulation of genetic damage (IEA) | GO:0007570
- obsolete aging (IEA) | GO:0007568
- obsolete extracellular carbohydrate transport (IEA) | GO:0006859
- obsolete negative regulation of cytochrome-c oxidase activity (IEA) | GO:1905376
- obsolete regulation of histone H3-K27 methylation (IEA) | GO:0061085
- obsolete regulation of histone H3-K79 methylation (IEA) | GO:2001160
- obsolete reproduction (IEA) | GO:0000003
- obsolete synthesis of RNA primer involved in premeiotic DNA replication (IEA) | GO:1902980
- ommochrome biosynthetic process (IEA) | GO:0006727
- phagocytosis (IEA) | GO:0006909
- plant-type cell wall modification (IEA) | GO:0009827
- positive regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902643
- positive regulation of antigen processing and presentation of peptide antigen via MHC class Ib (IEA) | GO:0002597
- positive regulation of dendrite extension (IEA) | GO:1903861
- positive regulation of double-strand break repair via nonhomologous end joining (IEA) | GO:2001034
- positive regulation of forebrain neuron differentiation (IEA) | GO:2000979
- positive regulation of oligopeptide transport (IEA) | GO:2000878
- positive regulation of oxidative phosphorylation (IEA) | GO:1903862
- positive regulation of phospholipid translocation (IEA) | GO:0061092
- positive regulation of proteasomal protein catabolic process (IEA) | GO:1901800
- positive regulation of response to water deprivation (IEA) | GO:1902584
- primary lung bud formation (IEA) | GO:0060431
- programmed cell death (IEA) | GO:0012501
- pronephric glomerulus morphogenesis (IEA) | GO:0035775
- pronuclear fusion (IEA) | GO:0007344
- propan-2-ol biosynthetic process (IEA) | GO:1902640
- protein localization to old growing cell tip (IEA) | GO:1903858
- pyrimidine nucleotide salvage (IEA) | GO:0032262
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate biosynthetic process (IEA) | GO:1902646
- regulation of 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate catabolic process (IEA) | GO:1902641
- regulation of antigen processing and presentation of lipid antigen via MHC class Ib (IEA) | GO:0002598
- regulation of cellular glucuronidation (IEA) | GO:2001029
- regulation of complement-dependent cytotoxicity (IEA) | GO:1903659
- regulation of cytokinin dehydrogenase activity (IEA) | GO:1903856
- regulation of ecdysteroid secretion (IEA) | GO:0007555
- regulation of endopeptidase activity (IEA) | GO:0052548
- regulation of gastric motility (IEA) | GO:1905333
- regulation of germ cell proliferation (IEA) | GO:1905936
- regulation of hemicellulose catabolic process (IEA) | GO:2000988
- regulation of intestinal epithelial cell development (IEA) | GO:1905298
- regulation of microglial cell migration (IEA) | GO:1904139
- regulation of phosphatidylcholine biosynthetic process (IEA) | GO:2001245
- regulation of phosphorus utilization (IEA) | GO:0006795
- regulation of plant epidermal cell differentiation (IEA) | GO:1903888
- regulation of protein maturation by peptide bond cleavage (IEA) | GO:0010953
- regulation of RNA binding transcription factor activity (IEA) | GO:1905255
- regulation of satellite cell activation involved in skeletal muscle regeneration (IEA) | GO:0014717
- regulation of skeletal muscle contraction by chemo-mechanical energy conversion (IEA) | GO:0014862
- response to rest involved in regulation of muscle adaptation (IEA) | GO:0014893
- retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum (IEA) | GO:0006890
- sesquiterpenoid biosynthetic process (IEA) | GO:0016106
- sophorose transport (IEA) | GO:2001087
- sperm aster formation (IEA) | GO:0035044
- sperm DNA decondensation (IEA) | GO:0035041
- synaptic vesicle docking (IEA) | GO:0016081
- synaptic vesicle priming (IEA) | GO:0016082
- tertiary alcohol biosynthetic process (IEA) | GO:1902645
- transcription by RNA polymerase V (IEA) | GO:0001060
- triterpenoid catabolic process (IEA) | GO:0016105
- tRNA import into mitochondrion (IEA) | GO:0016031
- xylotriose transport (IEA) | GO:2001094
 Domains
Domains
- ( PF00385 ) Chromo (CHRromatin Organisation MOdifier) domain
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
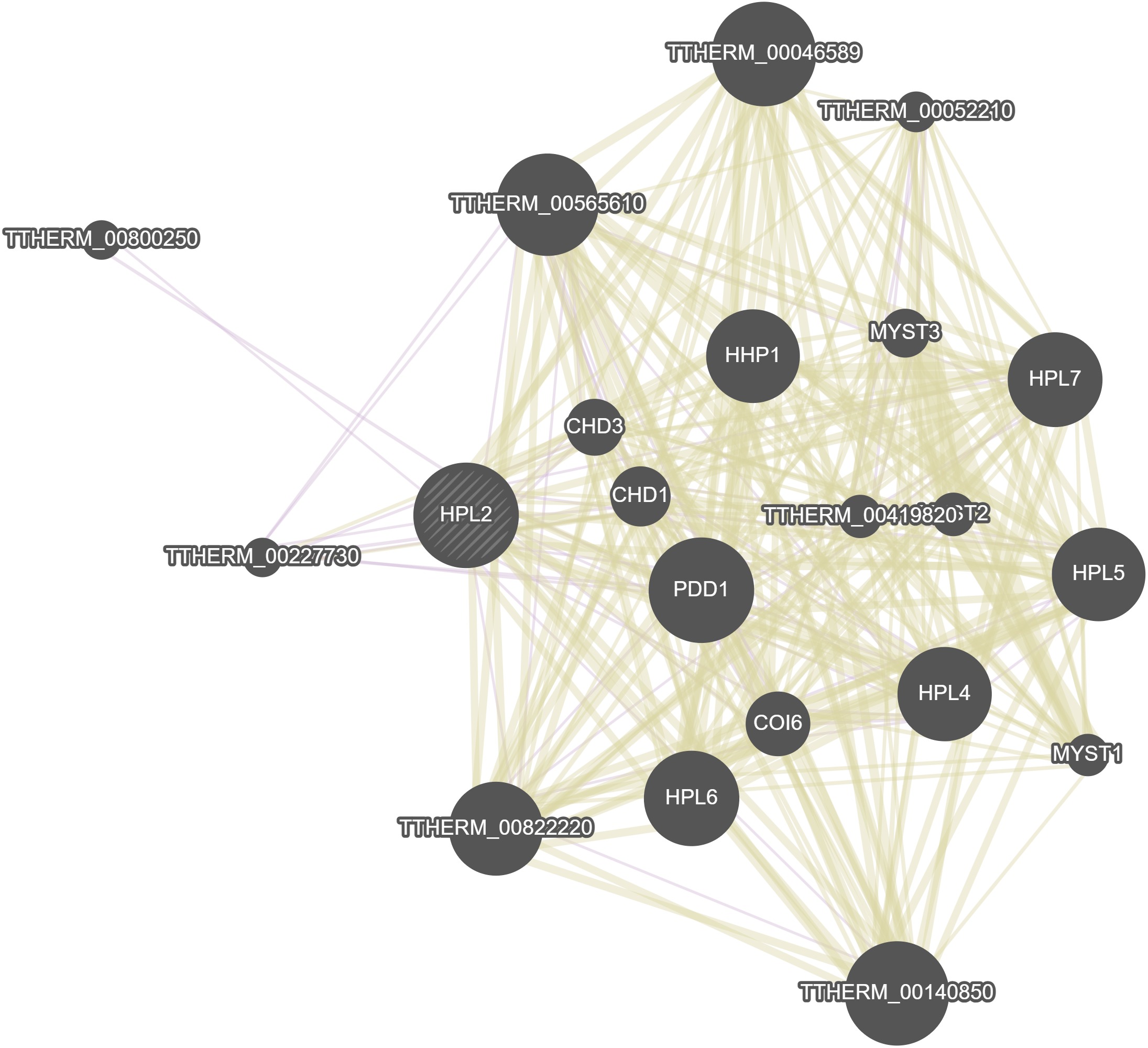
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2317 JUB1, JUB2, COI6, COI3, JUB4, HPL2 This protein co-localizes with Pdd1p in heterochromatin body in the new macronucleus.
 Associated Literature
Associated Literature
- Ref:28692189: Wiley EA, Horrell S, Yoshino A, C Schornak C, Bagnani C, L Chalker D (2017) Diversification of HP1-like Chromo Domain Proteins in Tetrahymena thermophila. The Journal of eukaryotic microbiology ( ):
- Ref:26688337: Kataoka K, Mochizuki K (2015) Phosphorylation of an HP1-like Protein Regulates Heterochromatin Body Assembly for DNA Elimination. Developmental cell 35(6):775-88
- Ref:25989344: Xu J, Yuan Y, Liang A, Wang W (2015) Chromodomain protein Tcd1 is required for macronuclear genome rearrangement and repair in Tetrahymena. Scientific reports 5( ):10243
 Sequences
Sequences
>TTHERM_01337400(coding)
ATGTTAATTAAATTGAATTTCAGTTTAGTATTTATCACGCTAATTATTAGCTTAGTTATT
GTTTATATTAGCTAAACTCGTAAACTTAATAAATAAAAATTTTTCTTAATTAATTACATA
AATTAAAAAAAACCTTTTGTGTAGATGTTCACTGTAAAGCAACAGGCACTAAATAAAATT
TAAAGTAAGTAGGCTTAACTTCCTAAAAATACATAAGATTCCCTTCAATAAATGCTTGCT
TGGGATTAAGAGTTTCTTGCATCAAAGGATAAGCTGTTTAAAAATAAGCATTAATACTAA
GGGAACGGTGGTTCAGCTTTGCTATTGAATTTAAACACTAACAAAAAGACAAATTTATAA
CAACAGTAACAACAGTAGAATTAAATCTAACCAACATAAATGAGCTAAATTTCGTAACAT
GAACTAAGGTAGTTGTAAAAAAAAAGAAAATTGCAAGAATTAGCAAGAGAAAATAGTAAA
ATTTTAAAGTCATAAAAGTAGCAATAAAAAGAAAATAATGAGTAAATAGATTCATCTTAG
GACGCCTCTGACGAATATGAAGTTGAAAAAATTGTTGATAAAAAAATTGAAAATGGATAA
ATATTTTACAAAGTGAAGTGGAAAGGTTGGGATAGTACTTACAATACTTGGGAACCTGAA
AATAATCTTTTTAGAGTATCTGAAATGATTGAAGAGTACGAAGCTTCAAAAAAATATTAA
AAAAGATAAAATTTCCCAAAATAAGTAGATTCTGGGAGTAGTGATTTGGAAGATAACGAT
AAAAAGTAAAATTCAGCTCCAAAAAAAGGAGAAACTTCAAATTAAATAAATTAAAGTAAC
CTATAAAATAAAAATGAGAGTTTGTCAAAACTTTAGTAGAGTTAGTAAAAAATTTAATCT
AATGTAATAAAAAGTAGATAGCCTTCTAATAATACACCAATAACCCCAAAAGAATAACAA
GATAGACTTTATAAAGAAAATTTAACTGCAAATAAATAGAATAATGCAAAAGAAATGGAT
ACATACGAAAAGAATTAAAATATGAGCAATAGCTAAATTTAACTTGAAAAAAATAAATAA
TTTTATTAATTGCTGATGAATTAAAAATCATAATCTGCTTCTTCTACTTCTGCTTTAATT
CCACCACAAAAACAGAATTAAAAGAATTTAGAAGAAACATAAAATTTGTAAAACAATATA
AAAAATTAAGAAAAAAAGATTGCTAACAACATAGAATAATTAGATCTAGAGGATGATAAT
CTTGAAGTTGAAAAAGATTATTCTTAGTAAATTCATCAAAAGTCACAAAACAAAGAAAAA
TCATTTGAAGAAAATGAAGATGGAAATGAGGACGACAAAGAATACATTTTTGAAGCCATA
CTTGATAAGCGTTAATTAGAGGGATAAGAAGTCGAATATTTGCTGAAATTTTCTAACCAT
GATAAACCTGAGTGGGAACTGATAAGTAATTTAGAAAGCATAGAAGAAGAAATAAAAGAA
TATGAAGAAGGGTTAAAAAGGGCTTAAGAAAAAGCTATTTTGCTATCATAATAAGAAAAT
TTAGATTAATAAAACTTGTAATTTGAAGAGGTTTAATAAGACAAAGAATTTATTAGTTAA
ACTGAACCTGCTACAAGTTTAACAACTATGCAAAGTCTAGTTCATTAAAATAAAGAAAAT
TAATCTTAAGTAAATCATCAGACTAAAAAAAATAAGATTGTCCCTGAATAAATAGATGTG
GACTGCTCAGATGATTAATTTGAACAAGAGGTTTAAAAAAATTAAATGGATATTTAATCT
AATTTAAGCCCATAAAATTCTAACCAAAATATCAATTATTAACCAGACTAATAGAAAAAA
GATAAATAAACTATTGGCTCTGAATTGAATAGTAAATAAAAATAGTAATAATTAAATATC
CAGCAAGTACTTCCTTGCTAAACTAAAATATTAGAAAATGTAATCTAAAATTAATAACCA
TTTAATTTAATTAATCCTTTACTTAATAACACTTCTCTTCAGCAGCTAGCAAATATTTCA
AACCCTATTAATTACACATAAAATTATAACTTTAGCTAGGAAATTGGATGCTTTTATAAA
GGTGATCAAAGATTTTCTATTGAACCTTGTGACTAATCTACTCTACAAGATCTTTCTAAG
ACTACTTTCAAAGTTATGTGGAAACCAAGAGTTAATGGTTGTGTACCTCTACCAAGTCTA
ATATCTTACTAAATGCTAAAAGAACATGATCCAGTAGGCTTGGTTTTATTTTTTGAAAAT
CTAATATAGAAATAATAGTAATAAATAGCTAAATGA>TTHERM_01337400(gene)
TACTCTATATATTAATTTGTTAACAAAAAATACTTCAATACTAAAGCTAGCCAACGATAA
CCCTAAAGTGCTTTTATTCAAGATAAACTTAATAATCTGAGGTACTTTTATTAGAGAAGC
TACTAAAGCAATTCCGATGCCAACTATTTTGGAGAAAAACTTTATTTTTAAATTAGATAT
TTAATAATAAAAATGAATTTAAAATTATCATATTTTTTTACTATGTTTAGACACACTTTG
TCATCAAAAAAGTTAGAAATTTCTGCATTCTATAAATTTTACAGCAAAGAAGGATAACAA
GAATGACTAGAGTCGAGTAAAATGTCTAGACCAGATTGAATTAAGCTCATTAATTTAATT
TGCAATCTTTTTATAGTTTATTTTTATCTAAATTCTTTTAATTTGAATTATTATTTACAA
ATTATTAGAGTGGTTTTTAGTAGTTTTGTAGATAGGGTGTGAACCAAAATGTTAATTAAA
TTGAATTTCAGTTTAGTATTTATCACGCTAATTATTAGCTTAGTTATTGTTTATATTAGC
TAAACTCGTAAACTTAATAAATAAAAATTTTTCTTAATTAATTACATAAATTAAAAAAAA
CCTTTTGTGTAGATGTTCACTGTAAAGCAACAGGCACTAAATAAAATTTAAAGTAAGTAG
GCTTAACTTCCTAAAAATACATAAGATTCCCTTCAATAAATGCTTGCTTGGGATTAAGAG
TTTCTTGCATCAAAGGATAAGCTGTTTAAAAATAAGCATTAATACTAAGGGAACGGTGGT
TCAGCTTTGCTATTGAATTTAAACACTAACAAAAAGACAAATTTATAACAACAGTAACAA
CAGTAGAATTAAATCTAACCAACATAAATGAGCTAAATTTCGTAACATGAACTAAGGTAG
TTGTAAAAAAAAAGAAAATTGCAAGAATTAGCAAGAGAAAATAGTAAAATTTTAAAGTCA
TAAAAGTAGCAATAAAAAGAAAATAATGAGTAAATAGATTCATCTTAGGACGCCTCTGAC
GAATATGAAGTTGAAAAAATTGTTGATAAAAAAATTGAAAATGGATAAATATTTTACAAA
GTGAAGTGGAAAGGTTGGGATAGTACTTACAATACTTGGGAACCTGAAAATAATCTTTTT
AGAGTATCTGAAATGATTGAAGAGTACGAAGCTTCAAAAAAATATTAAAAAAGATAAAAT
TTCCCAAAATAAGTAGATTCTGGGAGTAGTGATTTGGAAGATAACGATAAAAAGTAAAAT
TCAGCTCCAAAAAAAGGAGAAACTTCAAATTAAATAAATTAAAGTAACCTATAAAATAAA
AATGAGAGTTTGTCAAAACTTTAGTAGAGTTAGTAAAAAATTTAATCTAATGTAATAAAA
AGTAGATAGCCTTCTAATAATACACCAATAACCCCAAAAGAATAACAAGATAGACTTTAT
AAAGAAAATTTAACTGCAAATAAATAGAATAATGCAAAAGAAATGGATACATACGAAAAG
AATTAAAATATGAGCAATAGCTAAATTTAACTTGAAAAAAATAAATAATTTTATTAATTG
CTGATGAATTAAAAATCATAATCTGCTTCTTCTACTTCTGCTTTAATTCCACCACAAAAA
CAGAATTAAAAGAATTTAGAAGAAACATAAAATTTGTAAAACAATATAAAAAATTAAGAA
AAAAAGATTGCTAACAACATAGAATAATTAGATCTAGAGGATGATAATCTTGAAGTTGAA
AAAGATTATTCTTAGTAAATTCATCAAAAGTCACAAAACAAAGAAAAATCATTTGAAGAA
AATGAAGATGGAAATGAGGACGACAAAGAATACATTTTTGAAGCCATACTTGATAAGCGT
TAATTAGAGGGATAAGAAGTCGAATATTTGCTGAAATTTTCTAACCATGATAAACCTGAG
TGGGAACTGATAAGTAATTTAGAAAGCATAGAAGAAGAAATAAAAGAATATGAAGAAGGG
TTAAAAAGGGCTTAAGAAAAAGCTATTTTGCTATCATAATAAGAAAATTTAGATTAATAA
AACTTGTAATTTGAAGAGGTTTAATAAGACAAAGAATTTATTAGTTAAACTGAACCTGCT
ACAAGTTTAACAACTATGCAAAGTCTAGTTCATTAAAATAAAGAAAATTAATCTTAAGTA
AATCATCAGACTAAAAAAAATAAGATTGTCCCTGAATAAATAGATGTGGACTGCTCAGAT
GATTAATTTGAACAAGAGGTTTAAAAAAATTAAATGGATATTTAATCTAATTTAAGCCCA
TAAAATTCTAACCAAAATATCAATTATTAACCAGACTAATAGAAAAAAGATAAATAAACT
ATTGGCTCTGAATTGAATAGTAAATAAAAATAGTAATAATTAAATATCCAGCAAGTACTT
CCTTGCTAAACTAAAATATTAGAAAATGTAATCTAAAATTAATAACCATTTAATTTAATT
AATCCTTTACTTAATAACACTTCTCTTCAGCAGCTAGCAAATATTTCAAACCCTATTAAT
TACACATAAAATTATAACTTTAGCTAGGAAATTGGATGCTTTTATAAAGGTGATCAAAGA
TTTTCTATTGAACCTTGTGACTAATCTACTCTACAAGATCTTTCTAAGACTACTTTCAAA
GTTATGTGGAAACCAAGAGTTAATGGTTGTGTACCTCTACCAAGTCTAATATCTTACTAA
ATGCTAAAAGAACATGATCCAGTAGGCTTGGTTTTATTTTTTGAAAATCTAATATAGAAA
TAATAGTAATAAATAGCTAAATGAATTTTGTTTTGGTTGGTTGGCCTACTTAATTTAAAT
TGATTCAATAATAAATAATTAAATATAGCTTTATTAGTATTTATTTTAGCAAAATTATAT
TTTTGATGAATTCAAAATTTTATTTTATATTATTAGTTTC>TTHERM_01337400(protein)
MLIKLNFSLVFITLIISLVIVYISQTRKLNKQKFFLINYINQKKPFVQMFTVKQQALNKI
QSKQAQLPKNTQDSLQQMLAWDQEFLASKDKLFKNKHQYQGNGGSALLLNLNTNKKTNLQ
QQQQQQNQIQPTQMSQISQHELRQLQKKRKLQELARENSKILKSQKQQQKENNEQIDSSQ
DASDEYEVEKIVDKKIENGQIFYKVKWKGWDSTYNTWEPENNLFRVSEMIEEYEASKKYQ
KRQNFPKQVDSGSSDLEDNDKKQNSAPKKGETSNQINQSNLQNKNESLSKLQQSQQKIQS
NVIKSRQPSNNTPITPKEQQDRLYKENLTANKQNNAKEMDTYEKNQNMSNSQIQLEKNKQ
FYQLLMNQKSQSASSTSALIPPQKQNQKNLEETQNLQNNIKNQEKKIANNIEQLDLEDDN
LEVEKDYSQQIHQKSQNKEKSFEENEDGNEDDKEYIFEAILDKRQLEGQEVEYLLKFSNH
DKPEWELISNLESIEEEIKEYEEGLKRAQEKAILLSQQENLDQQNLQFEEVQQDKEFISQ
TEPATSLTTMQSLVHQNKENQSQVNHQTKKNKIVPEQIDVDCSDDQFEQEVQKNQMDIQS
NLSPQNSNQNINYQPDQQKKDKQTIGSELNSKQKQQQLNIQQVLPCQTKILENVIQNQQP
FNLINPLLNNTSLQQLANISNPINYTQNYNFSQEIGCFYKGDQRFSIEPCDQSTLQDLSK
TTFKVMWKPRVNGCVPLPSLISYQMLKEHDPVGLVLFFENLIQKQQQQIAK