Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00300660Standard Name
MND1 (Homolog of budding yeast MND1 (meiotic non disjunction)and fission yeast MCP7)Aliases
PreTt19361 | 28.m00320 | 3691.m00138Description
MND1 transmembrane protein putative; In budding yeast, the Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic DSB repair; Mnd1 requires Hop2 to localize to chromosomes; hypothetical proteinGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
No Data fetched for Gene Ontology Annotations
 Domains
Domains
No Data fetched for Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
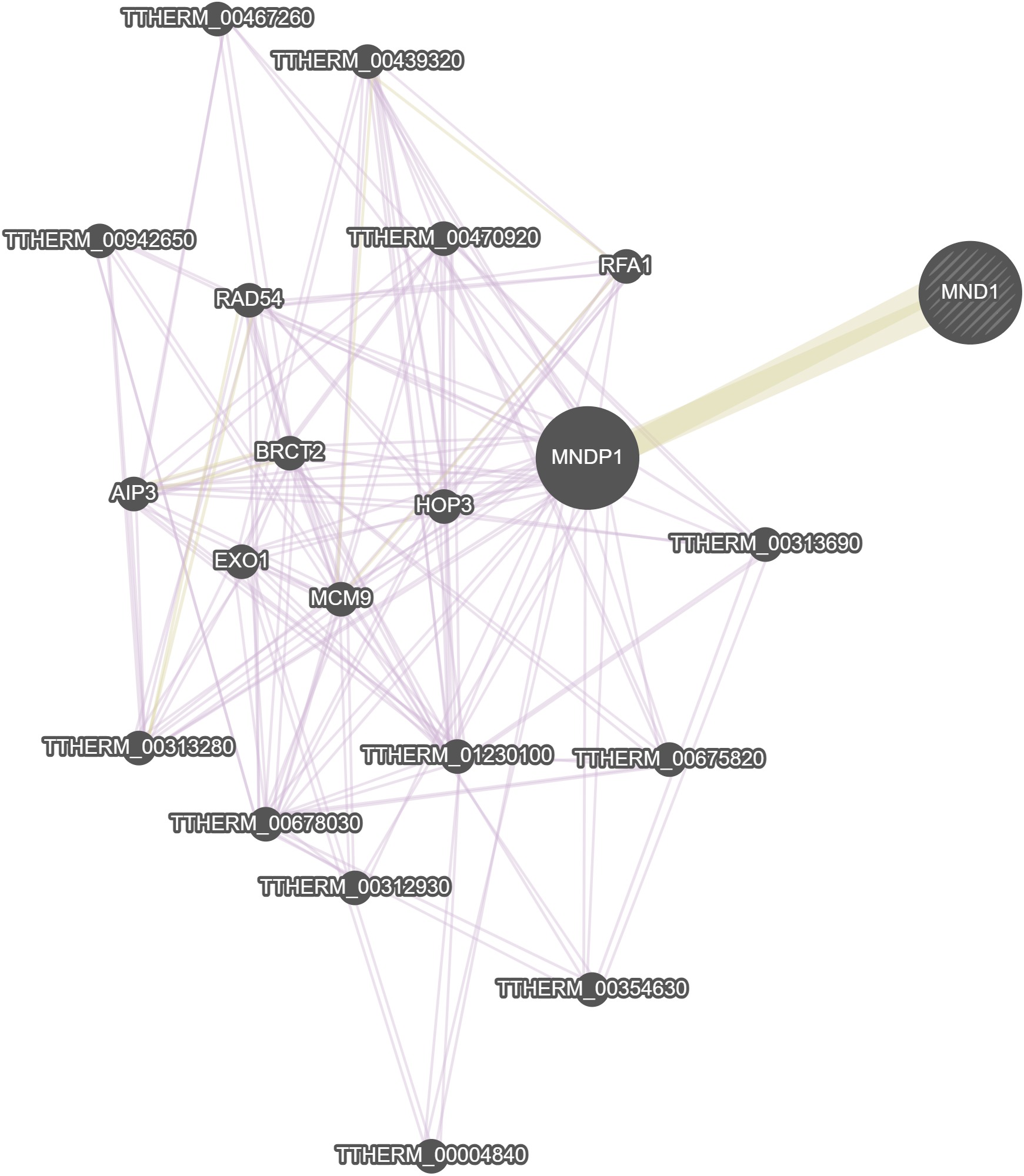
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2260 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists. 2261 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists. 2262 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists. 2263 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists. 2345 MND1 The gene model is incorrect. See gene_000003690 in TFGD for the transcript sequence
 Associated Literature
Associated Literature
- Ref:18522989: Mochizuki K, Novatchkova M, Loidl J (2008) DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. Journal of cell science 121(Pt 13):2148-58
 Sequences
Sequences
>TTHERM_00300660(coding)
ATGGGAAAAAGATTGGATAGATTTTTAAATTTTATAAAATAGATTGATAAAATAGATATT
TATGGCTAGAAGATTAGCCTCAAATTCATGAAAGAGTCTGAATATAAAACAAGGTTTGGA
GGCTTAATAACTTTAGTAGTTGGGTTATTGACTGGACTTTGGACCTTAGTTTCAATTTAT
CAGTTAATATATTCTTCTGTTAATTCAATCATATCTCAAGAATATGTAATGGATGCTTAA
GAATTAAATTCAATATCTTTTAATCCTAAGTCTTAAAATACATTTTAGGTAGGGTTTTTC
AATAAATAAACTCAATTGCCAATTGATTAGAATTATTTTAAAATAGTTTCATATTATAAT
TAAGAATGTGATATCATTAATTAAAAAGTAGAAGTATTATCTTTTTAGGAAATTCAAAGT
AAATCAGTGGAACTAACTGATTGCTAAGATCTTATAGAATCATCTTAAGAATATCAGTTT
TTAAGTTAAACAAAAGATGGGTATTTTTGCTTCAACAAAGACAAATCATTTACAGAAAAT
GAAGTTTATCCTGAAATTTAATAAACAGTTAACTGTCGTAATAGTATTAATATTTTGGTA
ACAAAGAGTAAAGATTTAGATGACTAAATTTTGTAAAACTTAAAGTAAAACAGTACTTTA
AGCTTGTCTGTTCCTACAACGTTAGTTGACTATACAGACATCCAAAATCCTTTCGATTAC
ACAAATATTATGAAACAATTTGACACAAATTTGTTTTTTAGTACTTAATCTAATATTTAA
TAAAAATTAATGATTTAAATAAATTTCTAAGCATTAATAATAGAAGATGAAAGAAATTAT
TCAATTCAATAAAATAGACTGATGAAATCTTTCGCAATATAATCAATTTCAGATAAACTT
ATCAATAGTGCTCTTAATCCTTAGGAGTAATCTTAGCTTAAAAATACCATTAACGATTCT
GATGTCTTATTCAAAATTAAGATTAGTTTTTCTGAATAAAACTACAATAAAAAATATATT
AGATCGTTTTAAAGCATACCTTCCATTCTAGGTAGTCTAGGAGGTGGTATTAAAATAGTA
GTTCTTATGGGATTTCTTATAGTTAGTCCTCTGTGCCAGATTCATTTTTAAAAGACTATG
CTAAATGAGGTGTTTAATTTTGAAAATGACTAAGATGATGATGATGATGAAAATTACTAA
AATAACTTTAATCTTCAGTCACTAAAATCTTTAGATGGGATATAAACAAATAACAAAAAT
CATATAAGTATTAATAACAACAGCAATATTTCTTAAGCCTAGAATTAAGATAATCCCAAA
AAATAGCAAGTCTCTAAAGGTTTATAGAAGATTAACAACTTTGAAAACTAATAAAATATA
TTGCAAACTAGTTCTCCAAAATATATTTAGGCTCTCAAAAATGTCACTAGTTTTATTTAA
ATTCAAAATATTAACTAAAAGTAAGAAAACTTACCCACTTCTACTAAAAGTATTACAAAT
TTTAAACAAATTAAAAATATAGAAAAGCCAATATAAAAACAAATCAATTAAATAACTAAT
AATTACACACATTAAAATAGCATGATATTTTCAAAGCATTCTCTTTAACAAGAAAACAAT
AATGATGAACTAAATTTTACGTAGCTAAGCTAACTGAAAGGTGGAGAAAGGAAAAACAAA
ATGAAAGAAATAATAAAGGGAACTATTTTTATCAATTCTTAAAGATAAATAGACTCTTAA
CAGTGTATAGATCAAAATTCCCCACTCCCTAAATTTCCTTAAAGATTATCATTTCTATCA
AACGCTTAATCATAGAAAACTATTGCCTTAACCAAAAAGAAATAAAATGACCAAAAGAAA
GCTGAAGAATAAATATTTGAAAAATATTTCAAGACAACATTCAATCCTCTAAAAATATCT
TTTATCGAAAGTTTATTTTACTATATATGGCCTTTCGGTAAATTCAAAAGAACCAAGGAA
CAGATGAGCTATTCTATTGATAAGCTAAATTACCATTTAGATAGTATTTACATCATTAAA
AAGCTACTTGAGATTGATAAGCTTAAAATGCTTTTATTAGATGAAAATTAAATTAAATTA
CTAGAATTCCTTCCAAAACCAACAATAAATTTGAAAAAAGTAGTAAACTAATAAGAATCT
TTTTAGGTTAGCTCTAATTTAATAACTGAAGATGATTAAAAAAAGATAAATATTTTGTAT
TAAGATACCAGAAATGAGTTAACAAAAGCTACACAAGCATTCTAAGCTTTCTAATCAATA
AAAGCCAAATCATCATTAACATAAATCGATAGAAAATTAATAGATTTACTGGATCCTAAA
ATTGCAGATTATTTTAATTAAGACAGTGATGATAATAAATCTTAAATAATTTAGTTAAAC
TAAATGAGTTTAGAAAACGAAAATAATCAACTTAATATCAATTATGATAAGGATACATCA
AATAAAATTTAACTTAATTATCAGTAAGAAAATACTACTCCTTAATCTTAGAATCTGATA
AAAGAAATTTACTAAAAAAGCAAAGGAAGCAGCGGTTATATTAGTTCTCCCTTGGAGATA
TACAGAAGTACAGCTAAGAAAGCATCTATGTTTAAATAACCACATATAAAGAGAGGATCT
CTTCATGTAACTTCTCAATATAATAGATAAGACGAAGTAGAAGAATATCAAAAAAATAAT
TAAATTTAGATAAAATATATACAAAATGATGGTTAAAGTGATTAGCATATACCTACAAGA
CTTACTCGCTTAAAATCATACTCAAGTGAAGATATTAATAAAATGCAAAGGCGAAGCAAG
GAAAGCTCGGTATCTAGCTAATAAAGTAAAGAAGGATCTAGCTTTTATTTTGGAGTTAAG
AATATTTCAGCACAATACCCTAAATCACTAGAAATGTAATATGTACTAGACGATTCTCAT
TTGTAGTAAGATGTAATTAATGGAGATATGGAAGAATCTCAGTAAAAATAACAATAAAAT
TAAAAAATATCTTTGGAGTTGAACAATGAAATAAATCATAGCAACCCTCCTGGATTGAAA
AAAATCTTCTCAAGCTCCCTAATCAGTAGTTGTAGTCATAGCATTATTGCAGATGAATCA
ATAAAACTCTAAAGTATAGATTGCCAAAATTTATAAATGTTTACAAAATAAAATAAATTT
TGTAAGTCTTCAAATTTGATAGTATCATAAAATTAGAAATAAGCAGAAAATTAGTCTGAA
TAAATTAATACCAGCAGTGAATAGACATATTTTAGAGAGAAAAAAAATTCTTCAAATGAA
GAAAATAAATTAGTTTCCCAATATGAAATTCAAATAAATTTACATAAAAAAGTTTAAAGT
TAAGAAAATATGAGTTGA>TTHERM_00300660(gene)
GCTAATGGTTAAGAGCTTTAAATTATTATTTTAATTAGTTTAATTAGTTAGAAATATCAC
TCTAAATAAACATACATATATTCATACAAATTTATCTTAGTAAGAAATTAATTCATCTAA
AATAGCAAAGAACTAATGGGAAAAAGATTGGATAGATTTTTAAATTTTATAAAATAGATT
GATAAAATAGATATTTATGGCTAGAAGATTAGCCTCAAATTCATGAAAGAGTCTGAATAT
AAAACAAGGTTTGGAGGCTTAATAACTTTAGTAGTTGGGTTATTGACTGGACTTTGGACC
TTAGTTTCAATTTATCAGTTAATATATTCTTCTGTTAATTCAATCATATCTCAAGAATAT
GTAATGGATGCTTAAGAATTAAATTCAATATCTTTTAATCCTAAGTCTTAAAATACATTT
TAGGTAGGGTTTTTCAATAAATAAACTCAATTGCCAATTGATTAGAATTATTTTAAAATA
GTTTCATATTATAATTAAGAATGTGATATCATTAATTAAAAAGTAGAAGTATTATCTTTT
TAGGAAATTCAAAGTAAATCAGTGGAACTAACTGATTGCTAAGATCTTATAGAATCATCT
TAAGAATATCAGTTTTTAAGTTAAACAAAAGATGGGTATTTTTGCTTCAACAAAGACAAA
TCATTTACAGAAAATGAAGTTTATCCTGAAATTTAATAAACAGTTAACTGTCGTAATAGT
ATTAATATTTTGGTAACAAAGAGTAAAGATTTAGATGACTAAATTTTGTAAAACTTAAAG
TAAAACAGTACTTTAAGCTTGTCTGTTCCTACAACGTTAGTTGACTATACAGACATCCAA
AATCCTTTCGATTACACAAATATTATGAAACAATTTGACACAAATTTGTTTTTTAGTACT
TAATCTAATATTTAATAAAAATTAATGATTTAAATAAATTTCTAAGCATTAATAATAGAA
GATGAAAGAAATTATTCAATTCAATAAAATAGACTGATGAAATCTTTCGCAATATAATCA
ATTTCAGATAAACTTATCAATAGTGCTCTTAATCCTTAGGAGTAATCTTAGCTTAAAAAT
ACCATTAACGATTCTGATGTCTTATTCAAAATTAAGATTAGTTTTTCTGAATAAAACTAC
AATAAAAAATATATTAGATCGTTTTAAAGCATACCTTCCATTCTAGGTAGTCTAGGAGGT
GGTATTAAAATAGTAGTTCTTATGGGATTTCTTATAGTTAGTCCTCTGTGCCAGATTCAT
TTTTAAAAGACTATGCTAAATGAGGTGTTTAATTTTGAAAATGACTAAGATGATGATGAT
GATGAAAATTACTAAAATAACTTTAATCTTCAGTCACTAAAATCTTTAGATGGGATATAA
ACAAATAACAAAAATCATATAAGTATTAATAACAACAGCAATATTTCTTAAGCCTAGAAT
TAAGATAATCCCAAAAAATAGCAAGTCTCTAAAGGTTTATAGAAGATTAACAACTTTGAA
AACTAATAAAATATATTGCAAACTAGTTCTCCAAAATATATTTAGGCTCTCAAAAATGTC
ACTAGTTTTATTTAAATTCAAAATATTAACTAAAAGTAAGAAAACTTACCCACTTCTACT
AAAAGTATTACAAATTTTAAACAAATTAAAAATATAGAAAAGCCAATATAAAAACAAATC
AATTAAATAACTAATAATTACACACATTAAAATAGCATGATATTTTCAAAGCATTCTCTT
TAACAAGAAAACAATAATGATGAACTAAATTTTACGTAGCTAAGCTAACTGAAAGGTGGA
GAAAGGAAAAACAAAATGAAAGAAATAATAAAGGGAACTATTTTTATCAATTCTTAAAGA
TAAATAGACTCTTAACAGTGTATAGATCAAAATTCCCCACTCCCTAAATTTCCTTAAAGA
TTATCATTTCTATCAAACGCTTAATCATAGAAAACTATTGCCTTAACCAAAAAGAAATAA
AATGACCAAAAGAAAGCTGAAGAATAAATATTTGAAAAATATTTCAAGACAACATTCAAT
CCTCTAAAAATATCTTTTATCGAAAGTTTATTTTACTATATATGGCCTTTCGGTAAATTC
AAAAGAACCAAGGAACAGATGAGCTATTCTATTGATAAGCTAAATTACCATTTAGATAGT
ATTTACATCATTAAAAAGCTACTTGAGATTGATAAGCTTAAAATGCTTTTATTAGATGAA
AATTAAATTAAATTACTAGAATTCCTTCCAAAACCAACAATAAATTTGAAAAAAGTAGTA
AACTAATAAGAATCTTTTTAGGTTAGCTCTAATTTAATAACTGAAGATGATTAAAAAAAG
ATAAATATTTTGTATTAAGATACCAGAAATGAGTTAACAAAAGCTACACAAGCATTCTAA
GCTTTCTAATCAATAAAAGCCAAATCATCATTAACATAAATCGATAGAAAATTAATAGAT
TTACTGGATCCTAAAATTGCAGATTATTTTAATTAAGACAGTGATGATAATAAATCTTAA
ATAATTTAGTTAAACTAAATGAGTTTAGAAAACGAAAATAATCAACTTAATATCAATTAT
GATAAGGATACATCAAATAAAATTTAACTTAATTATCAGTAAGAAAATACTACTCCTTAA
TCTTAGAATCTGATAAAAGAAATTTACTAAAAAAGCAAAGGAAGCAGCGGTTATATTAGT
TCTCCCTTGGAGATATACAGAAGTACAGCTAAGAAAGCATCTATGTTTAAATAACCACAT
ATAAAGAGAGGATCTCTTCATGTAACTTCTCAATATAATAGATAAGACGAAGTAGAAGAA
TATCAAAAAAATAATTAAATTTAGATAAAATATATACAAAATGATGGTTAAAGTGATTAG
CATATACCTACAAGACTTACTCGCTTAAAATCATACTCAAGTGAAGATATTAATAAAATG
CAAAGGCGAAGCAAGGAAAGCTCGGTATCTAGCTAATAAAGTAAAGAAGGATCTAGCTTT
TATTTTGGAGTTAAGAATATTTCAGCACAATACCCTAAATCACTAGAAATGTAATATGTA
CTAGACGATTCTCATTTGTAGTAAGATGTAATTAATGGAGATATGGAAGAATCTCAGTAA
AAATAACAATAAAATTAAAAAATATCTTTGGAGTTGAACAATGAAATAAATCATAGCAAC
CCTCCTGGATTGAAAAAAATCTTCTCAAGCTCCCTAATCAGTAGTTGTAGTCATAGCATT
ATTGCAGATGAATCAATAAAACTCTAAAGTATAGATTGCCAAAATTTATAAATGTTTACA
AAATAAAATAAATTTTGTAAGTCTTCAAATTTGATAGTATCATAAAATTAGAAATAAGCA
GAAAATTAGTCTGAATAAATTAATACCAGCAGTGAATAGACATATTTTAGAGAGAAAAAA
AATTCTTCAAATGAAGAAAATAAATTAGTTTCCCAATATGAAATTCAAATAAATTTACAT
AAAAAAGTTTAAAGTTAAGAAAATATGAGTTGAGGTTTTAAAAATATACTTAAACAAAAA
TAAATCAAAATAAATAATACCATTAATTCAGCTAAAAAATGTATAATTTTTGTCATATTT
TACTGTATTCCTTTTAATTTA>TTHERM_00300660(protein)
MGKRLDRFLNFIKQIDKIDIYGQKISLKFMKESEYKTRFGGLITLVVGLLTGLWTLVSIY
QLIYSSVNSIISQEYVMDAQELNSISFNPKSQNTFQVGFFNKQTQLPIDQNYFKIVSYYN
QECDIINQKVEVLSFQEIQSKSVELTDCQDLIESSQEYQFLSQTKDGYFCFNKDKSFTEN
EVYPEIQQTVNCRNSINILVTKSKDLDDQILQNLKQNSTLSLSVPTTLVDYTDIQNPFDY
TNIMKQFDTNLFFSTQSNIQQKLMIQINFQALIIEDERNYSIQQNRLMKSFAIQSISDKL
INSALNPQEQSQLKNTINDSDVLFKIKISFSEQNYNKKYIRSFQSIPSILGSLGGGIKIV
VLMGFLIVSPLCQIHFQKTMLNEVFNFENDQDDDDDENYQNNFNLQSLKSLDGIQTNNKN
HISINNNSNISQAQNQDNPKKQQVSKGLQKINNFENQQNILQTSSPKYIQALKNVTSFIQ
IQNINQKQENLPTSTKSITNFKQIKNIEKPIQKQINQITNNYTHQNSMIFSKHSLQQENN
NDELNFTQLSQLKGGERKNKMKEIIKGTIFINSQRQIDSQQCIDQNSPLPKFPQRLSFLS
NAQSQKTIALTKKKQNDQKKAEEQIFEKYFKTTFNPLKISFIESLFYYIWPFGKFKRTKE
QMSYSIDKLNYHLDSIYIIKKLLEIDKLKMLLLDENQIKLLEFLPKPTINLKKVVNQQES
FQVSSNLITEDDQKKINILYQDTRNELTKATQAFQAFQSIKAKSSLTQIDRKLIDLLDPK
IADYFNQDSDDNKSQIIQLNQMSLENENNQLNINYDKDTSNKIQLNYQQENTTPQSQNLI
KEIYQKSKGSSGYISSPLEIYRSTAKKASMFKQPHIKRGSLHVTSQYNRQDEVEEYQKNN
QIQIKYIQNDGQSDQHIPTRLTRLKSYSSEDINKMQRRSKESSVSSQQSKEGSSFYFGVK
NISAQYPKSLEMQYVLDDSHLQQDVINGDMEESQQKQQQNQKISLELNNEINHSNPPGLK
KIFSSSLISSCSHSIIADESIKLQSIDCQNLQMFTKQNKFCKSSNLIVSQNQKQAENQSE
QINTSSEQTYFREKKNSSNEENKLVSQYEIQINLHKKVQSQENMS