Identifiers and Description
Identifiers and Description
Gene Model Identifier
TTHERM_00382290Standard Name
MNDP1 (Homolog of budding yeast MND1 (meiotic non disjunction)and fission yeast MCP7)Aliases
PreTt10386 | 40.m00271Description
MNDP1 Mnd1 family protein; In budding yeast, the Mnd1 protein forms a complex with Hop2 to promote homologous chromosome pairing and meiotic DSB repair; Mnd1 requires Hop2 to localize to chromosomes; There exists an ubiquitously expressed paralog, MNDP1; Leucine zipper with capping helix domainGenome Browser (Macronucleus)
Genome Browser (Micronucleus)
 External Links
External Links
 Gene Ontology Annotations
Gene Ontology Annotations
Biological Process
- meiosis I (ISA) | GO:0007127
 Domains
Domains
 Gene Expression Profile
Gene Expression Profile
 Vegetative Cell Cycle (Zhang et al.,
2023)
Vegetative Cell Cycle (Zhang et al.,
2023)
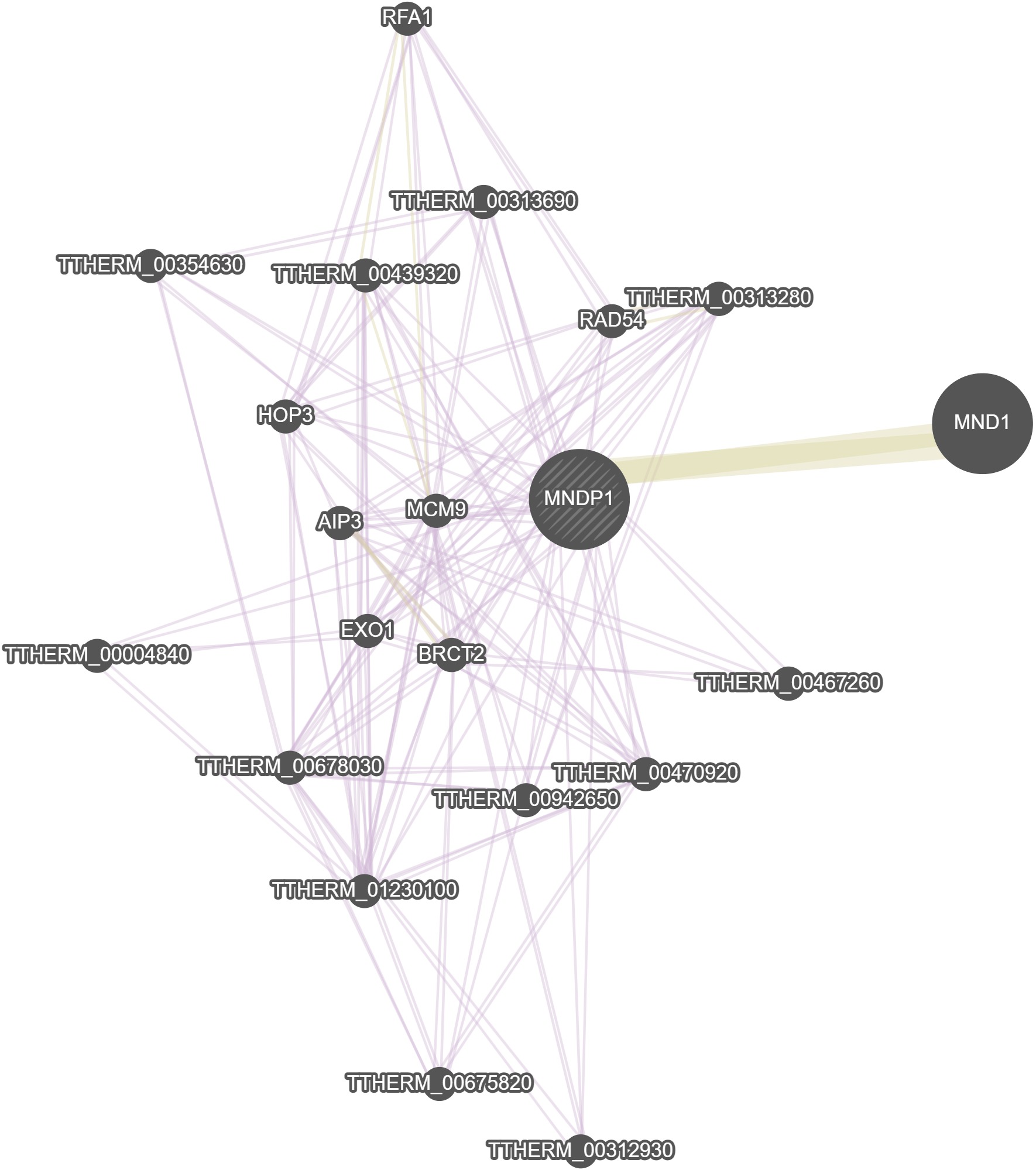
 GeneMania
GeneMania
 Tetrahymena Stock Center
Tetrahymena Stock Center
No Data fetched for Tetrahymena Stock Center
 Homologs
Homologs
No Data fetched for Homologs
 General Information
General Information
No. Gene Name(s) Paragraph Text 2260 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists. 2261 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists. 2262 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists. 2263 MND1, MNDP1, HOP3 Hop2 and Mnd1 are meiosis-specific proteins that function in a complex in budding yeast. Their general architecture is strikingly similar, and therefore they are potentially homologous protein families. The Hop2-Mnd1 system seems to have undergone duplication in the evolutionary history of Tetrahymena, because both protein families are represented by two homologs with distinct expression patterns in this species. Just as for HOP2 (meiotic) and HOPP2 (ubiquitous), there is a meiotic (MND1)and a ubiquitously expressed (MNDP1) version, which raises the possibility that a meiotic and a ubiquitous Mnd1p-Hop2p complex exists.
 Associated Literature
Associated Literature
- Ref:18522989: Mochizuki K, Novatchkova M, Loidl J (2008) DNA double-strand breaks, but not crossovers, are required for the reorganization of meiotic nuclei in Tetrahymena. Journal of cell science 121(Pt 13):2148-58
 Sequences
Sequences
>TTHERM_00382290(coding)
ATGGCTAAAAGAACTAAACCTTTAACTCTTGAATAGAAGAGAGAAAAAATGCTCCATGTC
TTCCATGAAACTCAATAAGTTTACAACTTGAAGGAAATTGAAAAATTCTCCACAAAGGCA
GGCATTGTACTTCCTACTGTAAAGGATGTTTTGATGAGTTTGATTGCAGATGGACTTGTT
GATTCTGATAAAATTGGTTCTGCTAACTTTTACTGGTCTCTCCCTTCACAAACTTACTTG
ATTTTAAAATTAAGATTAGCTGAAGCAGAACAAAAAATGAAAGATTGCGATTAAAAAAAG
GTAGAATTAGAGGATGAGCTAGAGAAATCCGTTATAGATAAAGAACCAAGCGATGATCGA
ACAGAATAGCTGAACAAATTAAAGGAATTGTAAGAAACTTTAAAGAAGAAGGAAAAGGAT
TTAGAAAAATACAAGAAATACGATCCTGAAAGACTAAAACAACTAGAGGAGTAAACTGAA
GATCTTAAAGCTAAGGTTAATCTGTGGACTGATAACTTATTCTCTTTAAAATCTTACATA
TAAAAATTAACTCCTATGGATGATGAAGAAGTTGAAAAAGCCTTCTCTATTCCAGCTGAT
CTCGATAATGTATGA>TTHERM_00382290(gene)
AAATACGCCAATATTGATTGTGTTTGAATTTAAGTACAATAGATACAATTTTCTGCAAAA
ATTTAATTATTATATTACAAAAAATAATCTAAATAGCTTTTGTAAAGTAAAATATTCATT
AGACACCACCAATTTATACCTAATCACTTTTAAGAAATGGCTAAAAGAACTAAACCTTTA
ACTCTTGAATAGAAGAGAGAAAAAATGCTCCATGTCTTCCATGAAACTCAATAAGTTTAC
AACTTGAAGGAAATTGAAAAATTCTCCACAAAGGCAGGCATTGTACTTCCTACTGTAAAG
GATGTTTTGATGAGTTTGATTGCAGATGGACTTGTTGATTCTGATAAAATTGGTTCTGCT
AACTTTTACTGGTCTCTCCCTTCACAAACTTACTTGATTTTAAAATTAAGATTAGCTGAA
GCAGAACAAAAAATGAAAGATTGCGATTAAAAAAAGGTAGAATTAGAGGATGAGCTAGAG
AAATCCGTTATAGATAAAGAACCAAGCGATGATCGAACAGAATAGCTGAACAAATTAAAG
GAATTGTAAGAAACTTTAAAGAAGAAGGAAAAGGATTTAGAAAAATACAAGAAATACGAT
CCTGAAAGACTAAAACAACTAGAGGAGTAAACTGAAGATCTTAAAGCTAAGGTTAATCTG
TGGACTGATAACTTATTCTCTTTAAAATCTTACATATAAAAATTAACTCCTATGGATGAT
GAAGAAGTTGAAAAAGCCTTCTCTATTCCAGCTGATCTCGATAATGTATGATAGTTGAAC
CAACAACTAAATAGTAAGCACTAACTAACTATCTACAAATTTTAAAGTGAAACACAAAAG
ATAAAATAAATTTATTCATACATTCATTCATTCATAATTATA>TTHERM_00382290(protein)
MAKRTKPLTLEQKREKMLHVFHETQQVYNLKEIEKFSTKAGIVLPTVKDVLMSLIADGLV
DSDKIGSANFYWSLPSQTYLILKLRLAEAEQKMKDCDQKKVELEDELEKSVIDKEPSDDR
TEQLNKLKELQETLKKKEKDLEKYKKYDPERLKQLEEQTEDLKAKVNLWTDNLFSLKSYI
QKLTPMDDEEVEKAFSIPADLDNV